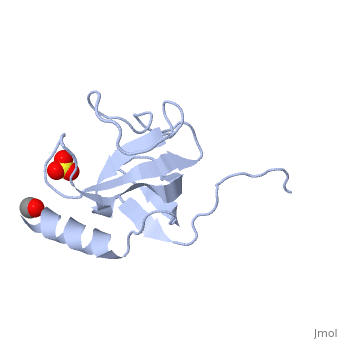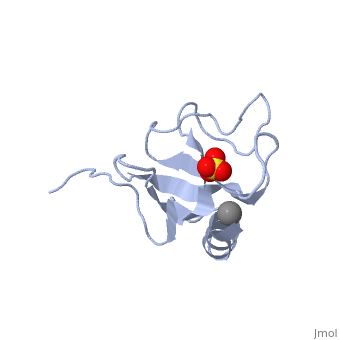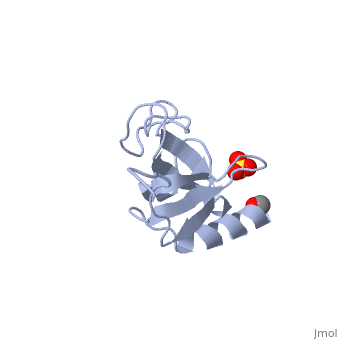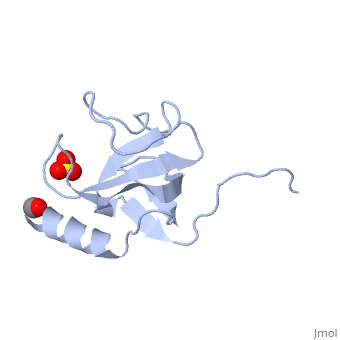
CAP-Gly domain

| |||||||||
| 1tov, resolution 1.77Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Gene: | F53F4.3 (Caenorhabditis elegans) | ||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
CAP-Gly domains in cytoskeletal proteinsCAP-Gly domains in cytoskeletal proteins
ActivityActivity
StructureStructure
PDB files 1tov and 1lpl[1] show the CAP-Gly domain from Caenorhabditis elegans. In the crystal, a dimer is formed by binding the C-terminus of one chain into the binding groove of the second chain.
1TOV is a rebuilt and re-refined version of the same dataset from 1LPL, giving an improvement of about 4% in Rfree, identifying a sulfate (seen in default images), and adding an additional turn of ordered helix toward the N-terminal end of the domain fragment.[2] Both are from the SouthEast Collaboratory for Structural Genomics (SECSG).
ReferencesReferences
- ↑ Li S, Finley J, Liu ZJ, Qiu SH, Chen H, Luan CH, Carson M, Tsao J, Johnson D, Lin G, Zhao J, Thomas W, Nagy LA, Sha B, DeLucas LJ, Wang BC, Luo M. Crystal structure of the cytoskeleton-associated protein glycine-rich (CAP-Gly) domain. J Biol Chem. 2002 Dec 13;277(50):48596-601. Epub 2002 Sep 7. PMID:12221106 doi:http://dx.doi.org/10.1074/jbc.M208512200
- ↑ Arendall WB 3rd, Tempel W, Richardson JS, Zhou W, Wang S, Davis IW, Liu ZJ, Rose JP, Carson WM, Luo M, Richardson DC, Wang BC. A test of enhancing model accuracy in high-throughput crystallography. J Struct Funct Genomics. 2005;6(1):1-11. PMID:15965733 doi:10.1007/s10969-005-3138-4