Insulin-Degrading Enzyme
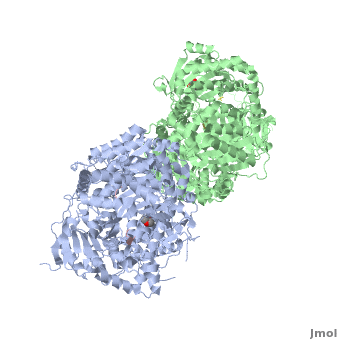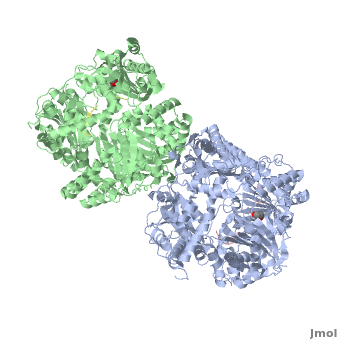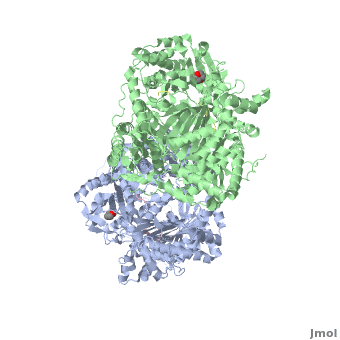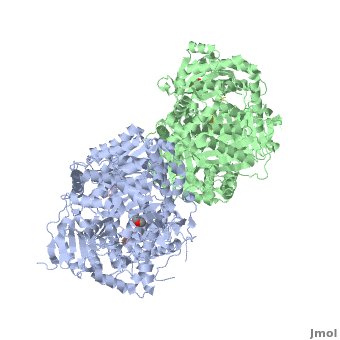
Insulin-Degrading Enzyme (PDB entry 2g49) is a highly conserved 2*113 kDa homodimeric zinc metalloprotease. It belongs to the M16A family of metalloproteases within the ME clan. It hydrolyses insulin and several amyloidogenic peptides like amyloid-β (Aβ).
Aβ is the main component of brain and of the amyloid islets found in Alzheimer’s disease.
Insulin is a peptide associated with diabete. The gene encoding IDE is located on human chromosome 10q and is in a region linked to type 2 diabete and Alzheimer’s disease.
The deletion of IDE in mice leads to hyperinsulinemia, glucose intolerance and increases cerebral Aβ accumulation. Conversely, enhancement of IDE activity in neurons significantly reduces Aβ levels in the brain.
IDE could help creating IDE-based therapies to control cerebral Aβ and blood sugar concentrations thanks to the understanding of its molecular mechanisms.
IDE is an unusual enzyme because of its high affinity for substrates that are widely diverse in sequence and structure. IDE prefers to degrade <6 kDa bioactive peptides such as insulin, Aβ, glucagon, atrial natriuretic peptides or transforming growth factor α. Paradoxally, even though IDE targets a broad range of substrates, it shows a remarkable capacity to selectively cleave some peptides without degrading related family members.
|
|
Structure general pointsStructure general points
IDE is a homodimeric protein. Each IDE monomer is composed of four 25-27 kDa structurally homologous αβ roll domains:
- : residues 43-285
- : residues 286-515
- : residues 542-768
- : residues 769-1,016
These domains share less than 25 % sequence similarity, and show similar secondary structures. The N-terminal domains 1 and 2 form a αβαβα sandwich (), as do domains 3 and 4 (). The size of each IDE-N and IDE-C is about 56 kDa. They are joined by a (residues 516-541) which interacts with both domains 2 and 3, but not with domains 1 and 4. There is an extensive interaction between IDE-N and IDE-C domains, that bury a large surface ( 11,200 Ȧ2), with good shape complementarity. This interaction between the two domains of IDE is mediated by charge complementarity and Van der Waals interactions. IDE-C is also involved in the oligomerization of IDE.
Catalytic chamberCatalytic chamber
IDE-N and IDE-C create substantial contact to form an enclosed catalytic chamber to encapsulate its substrates. This enclosed chamber is shaped like a triangular prism, with triangular base dimensions of 35x34x30 Ȧ and a height of 36 Ȧ. So the total volume of this prism is about 1.3x104 Ȧ3. All four domains contribute to surface the inner chamber. The surface provided by IDE-N is largely neutral or negatively charged, whereas that provided by IDE-C is mainly positively charged. The catalytic site of IDE is present within IDE-N, in the domain 1, but IDE-C is necessary for its catalytic activity. The contains a Zn2+ ion coordinated by two histidines (residues 108 and 112) and two glutamates (residues 111 and 189). The glutamate 111-residue is a base for the activation of a catalytic water molecule that mediates peptides hydrolysis. Moreover, these water molecules are involved in the coordination of the Zn2+ ion. At the catalytic site, these residues create a largely polar cavity with parts of hydrophobic and charged regions that interact with cleavage sites in many substrates. In the domain 2, there is an exosite that accommodates the N-terminus of substrates and is involved in the substrates binding.
Substrate bindingSubstrate binding
IDE can adopt two conformations, called “open” (IDE-o) and “closed” (IDE-c). In the open state, substrates can freely diffuse in and out of the catalytic chamber. In the close state, previously bound substrates are entrapped inside the catalytic chamber and repositioned to allow hydrolysis. The enclosed substrate-binding compartment of IDE prevents the entry and exit of substrates, so that substrates cannot be entrapped unless they were previously bound. IDE has to undergo a switch between these two states for catalysis. The enclosed substrate undergoes conformational changes to form β-sheets with two discrete regions of IDE for its degradation. IDE-N and IDE-C have extensive interactions that bury a large surface of 11,496 Ȧ2 with good shape complementarity and numerous hydrogen bonds. In the absence of interactions with factors or proteins, the substrate-free IDE-c state is stable and the catalytic chamber of IDE is mostly closed. So we can consider that IDE-c is the resting and inactive state of IDE.
3D structures of insulin-degrading enzyme3D structures of insulin-degrading enzyme
3n56, 3n57 – hIDE residues 42-1019 (mutant) + B-type natriuretic peptide – human
3ofi - hIDE residues 42-1019 + ubiquitin
3h44 - hIDE residues 42-1019 (mutant) + C-C motif chemokine 3
3hgz, 2g48 - hIDE residues 42-1011 (mutant) + amylin
3e4z - hIDE residues 42-1019 (mutant) + insulin-like growth factor II
3e50 - hIDE residues 42-1011 (mutant) + Transforming growth factor α
3e4a – hIDE (mutant) + hydroxamate peptide II1
2wby, 2wc0 - hIDE residues 42-1019 (mutant) + insulin
3cww - hIDE residues 42-1019 (mutant) + Bradykinin N terminal peptide
2jbu - hIDE residues 42-1019 (mutant) + CO-purified peptide
2g47 - hIDE residues 42-1019 (mutant) + amyloid β A4
2g49 - hIDE residues 42-1019 (mutant) + glucagons
2g54 - hIDE residues 42-1019 (mutant) + insulin β chain + Zn
ReferencesReferences
- ↑ Im H, Manolopoulou M, Malito E, Shen Y, Zhao J, Neant-Fery M, Sun CY, Meredith SC, Sisodia SS, Leissring MA, Tang WJ. Structure of substrate-free human insulin-degrading enzyme (IDE) and biophysical analysis of ATP-induced conformational switch of IDE. J Biol Chem. 2007 Aug 31;282(35):25453-63. Epub 2007 Jul 5. PMID:17613531 doi:10.1074/jbc.M701590200
- ↑ Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006 Oct 19;443(7113):870-4. Epub 2006 Oct 11. PMID:17051221 doi:10.1038/nature05143
- ↑ Li P, Kuo WL, Yousef M, Rosner MR, Tang WJ. The C-terminal domain of human insulin degrading enzyme is required for dimerization and substrate recognition. Biochem Biophys Res Commun. 2006 May 19;343(4):1032-7. Epub 2006 Mar 22. PMID:16574064 doi:10.1016/j.bbrc.2006.03.083