Cytochrome c
| |||||||||
| 3cp5, resolution 1.24Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Gene: | cytC (Rhodothermus marinus) | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
The cytochrome c (cyt c) proteins are a superfamily belonging to the class of all-α proteins, which are denoted as such by having an α-helical core. Each protein in this superfamily also contains one or more covalently-bound heme prosthetic groups.[1][2] The cyt c superfamily contains many different families, some of which are better characterized than others. These families include monodomain and multi-domain C-type cytochromes, such as cyt c4, a diheme C-type cytochrome, and NrfB, a pentaheme C-type cytochrome. In particular, the monoheme cyt c from Rhodothermus marinus has been previously studied and provides an excellent example of how some protein characteristics and structures can be extremely diverse, yet conserved, through evolution.
IntroductionIntroduction
Cytochromes are a class of heme-containing proteins found in bacteria and the mitochondria of eukaryotes.[2] These proteins are generally membrane-bound and are known as respiratory pigments because they are involved in various electron transport systems in oxidative phosphorylation.[3] Cytochromes can be categorized into several different types, three of which are based on the type of heme group the cytochrome contains: cytochromes a, b and d contain heme a, b and d, respectively.[4] Cytochrome c is named such because it contains the heme c, but is mainly distinguished from cytochromes a, b and d due to the heme being coordinated with the protein scaffold by cysteinyl residues covalently bound to either one or both of the heme's vinyl side chains.[3]
Cyt c has been split into four classes.[4] Class I contains soluble, low spin[2] single domain C-type cytochromes, of which there has been at least six subclasses found in prokaryotes including Desulfovibrio desulfuricans, Rhodospirillum rubrum, and Rhodothermus marinus. Cyt c in this class have a single heme attached close to the N-terminus of the polypeptide, with a methionine residue being the sixth iron coordination site. Class II contains higher spin-state cytochromes c, such as cyt c', with the heme being attached closer to the C-terminus. Class III contains cytochromes with multiple heme groups; these proteins have lower redox potentials compared to the other three classes[4]. Finally, Class IV is comprised of more complex proteins with higher molecular weights containing heme c as well as other prosthetic groups.[5]
Rhodothermus marinus cytochrome cRhodothermus marinus cytochrome c
StructureStructure
|
All members in the C-type cytochrome superfamily contain a heme prosthetic group that is covalently attached to the protein via two thioether bonds to cysteine residues. Most cytochromes c occur in a where the histidine residue is one of the two axial ligands of the heme iron.[2][3] In monoheme cytochromes c, the other axial position may be left vacant or be occupied by histidine or methionine residues; however, it can sometimes be occupied by cysteine or lysine residues.[2]. In Rmcytc, XX represents a threonine (Thr46) and an alanine residue (Ala47) that help form the loop 2 structure.
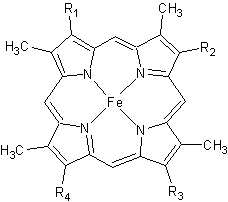
The typical monoheme cyt c fold is formed by helices . Rmcytc contains seven α-helices that are folded around the heme, all connected by random coils.[2] The heme group is axially coordinated by , and the disulfide linkages exist at . The heme group in Rmcytc is almost completely shielded from solvent due to it being in a mostly hydrophobic pocket. This pocket is formed in part by the seven helices surrounding the ring, but also by two structures that are uncommon in other cytochromes c. First, a 21 amino acid extension of the N-terminal exists, forming , which wraps around the back of the polypeptide.[2] An extension resembling such has only been seen in Thermus thermophilus; however, the extension occurs at the C-terminus rather than the N-terminus.[6] A second rarity is that of , inserted between helix D and loop 3, that shields the bottom part of the heme from any solvent.[2] In cytochrome c2 as well as mitochondrial cyt c, a similar yet shorter helix was found, though this helix was present at a different place in the primary sequence. Also, instead of helix B', T. thermophilus contains a two-stranded β-sheet.[2] One final note is the number of residues that Rmcytc contains. In general, cyt c contains about two methionines whereas Rmcytc contains seven, located on the left of the heme.[2]
As determined by X-ray crystallography, the Rmcytc structure was found to contain a sulfate ion coordinated to Glu122 via hydrogen bonding to the protonated carboxylate oxygen. In the protein complex, this ion has been seen to mediate crystal contact between neighbouring protein molecules.[2]
The observation of these structural motifs in other C-type cytochromes can support the divergent evolution of cytochromes c.[2] These motifs are present in a number of different bacteria and are seen in similar regions of the secondary structure; however, they exist in the primary sequence in places distinct to the phylum. For example, monoheme cytochromes c in the rest of the Bacteroidetes phylum have an N-terminus extension that is highly conserved to that of Rmcytc, and the regions in the primary structure that correspond to these secondary motifs are not observed in other bacterial phyla.[2] Also, due to these motifs being absent from other phyla, the Bacteroidetes monoheme cyt c has been said to form a new subfamily of cyt c.
FunctionFunction
Monoheme cytochromes c are involved in electron transport chains in both prokaryotes and eukaryotic mitochondria.[2] They mediate the transfer of electrons mainly from the bc1 complexes or their analogs to heme-copper oxygen reductases (HCOs) in the electron transport chain of oxidative phosphorylation. Heme c containing domains are often found fused to other protein domains such as these HCOs, including the caa3 oxygen reductases[2][7]; these enzymes are membrane-bound and catalyze the reduction of O2 to water.[8] In addition to being involved in oxidative phosphorylation, monoheme cyt c has also been seen to participate in the electron transport chain of photosynthesis.[2] Cytochrome c has also been determined to be a major signalling molecule in the apoptotic pathways.
Electron transport chainElectron transport chain
In the electron transport chain (ETC), cyt c shuttles electrons between the respiratory complexes III and IV; complex III is the cytochrome bc1 complex and IV is cyt c oxidase. Initially, the heme iron in cyt c is in the reduced, Fe3+ state; this allows for the uptake of one electron, oxidizing the iron to the Fe2+ state.[9] The ETC in eukaryotes is quite simple compared to that of prokaryotes (Figure 3).
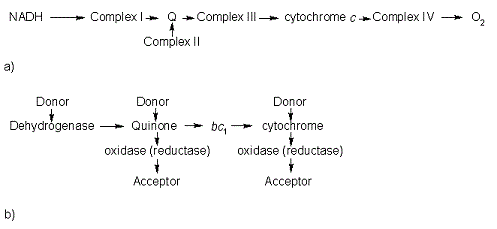
In prokaryotic systems, electrons can enter the ETC at a number of places and multiple donors can be in play; however, the underlying transport system remains the same. Electrons are ultimately transferred from donor to various redox complexes including the bc1 complex and cytochrome c, and finally to a terminal electron acceptor such as molecular oxygen in eukaryotes.[9]
The cytochrome oxidase reaction accounts for nearly 90% of all oxygen uptake in most cells.[9] Due to the large role of cytochromes within the ETC, it would be highly detrimental to the cell if any inhibitors were to be present in the organism. Cyanide and azide bind tightly to the cytochrome oxidase complex, halting electron transport and reducing the overall ATP production.[9]
ApoptosisApoptosis
In all organisms, cells undergo apoptosis, or programmed cell death, by which there is an extrinsic and an intrinsic pathway. The extrinsic pathway involves an immune response by killer lymphocytes, and once the lymphocyte has been bound to the target cell, an apoptotic cascade occurs.[9] The intrinsic pathway includes cyt c, present in the intermembrane space of mitochondria. In this pathway, the presence of an apoptotic stimulus causes cyt c to be released into the cytosol. Cytochrome c in the cytosol now can be recognized and bound to various apoptotic factors, activating them and forming the apoptosome. The apoptosome recruits caspases, which are activated and result in a caspase cascade to proceed with apoptosis.[9]
Cytochrome c is required for the intrinsic apoptotic process to function properly. Such as with the electron transport chain, a mutation affecting cyt c or other structures in apoptosis could cause either an increase or a decrease in the rate of apoptosis.
3D structures of cytochrome C3D structures of cytochrome C
Cytochrome CCytochrome C
3nwv – hCyt (mutant) – human
1j3s – hCyt - NMR
3nbs, 3nbt, 1crc, 1hrc – hoCyt – horse
1lc1, 1lc2, 1m60, 1giw, 2giw, 1akk, 2frc, 1ocd – hoCyt – NMR
1fi9 - hoCyt + imidazole – NMR
1u75 - hoCyt + Cyt peroxidase
1wej – hoCyt + Fab fragment
3a9f – Cyt C-terminal – Chlorobaculum tepidum
3cp5 – Cyt residues 29-152 – Rhodothermus marinus
2jti – yCyt (mutant) + Cyt peroxidase – yeast
2pcb - yCyt + Cyt peroxidase
2gb8 - yCyt + Cyt peroxidase - NMR
2jqr - yCyt (mutant) + adrenodoxin
2orl - yCyt (mutant) – NMR
1crg, 1crh, 1cri, 1crj, 2ycc - yCyt
1ytc, 1cie, 1cif, 1cig, 1cih, 1csu, 1csv, 1csw, 1csx, 1chh, 1chi, 1chj, 1cty, 1ctz - yCyt (mutant)
1rap, 1raq, 1ycc- yCyt iso-1
1yic – yCyt iso-1 – NMR
1irv, 1irw – yCyt iso-1 (mutant)
1fhb - yCyt iso-1 (mutant) + CN - NMR
1nmi – yCyt iso-1 + imidazole
2b0z, 2b10, 2b11, 2b12, 1u74, 1s6v – yCyt iso-1 (mutant) + Cyt peroxidase
2pcc – yCyt iso-1 + Cyt peroxidase
1yea, 1yeb – yCyt iso-2
2e84 – DvCyt – Desulfovibrio vulgaris
2j7a – DvCyt catalytic + electron donor subunits
2oz1 – Cyt – Rhodovulum sulfidophilum
2aiu – Cyt – mouse
2fw5, 2fwt – RsCyt diheme residues 1-139 - Rhodobacter sphaeroides
1dw0, 1dw3 - RsCyt diheme residues 1-112
1dw1, 1dw2 - RsCyt diheme residues 1-112 + small molecule
1ogy - RsCyt diheme residues 25-154 + nitrate reductase catalytic subunit
2a3m, 2a3p – DdCyt tetraheme membrane-bound subunit - Desulfovibrio desulfuricans
1h21 - DdCyt di-heme
1ofw, 1ofy, 1duw, 19hc - DdCyt nine-heme
1oah - DdCyt
2b4z – bCyt – bovine
1lfm, 1i55, 3cyt – Cyt – tuna
1fs7, 1fs8, 1fs9 – WsCyt + small molecule – Wolinella succinogenes
1dxr – RvCyt in photosynthetic reaction center – Rhodopseudomonas viridis
1qdb – Cyt – Sulfurospirillum deleyianum
5cyt – Cyt - albacore
Cytochrome C’Cytochrome C’
2xl6, 2xld, 2xle, 2xlo, 2xlv, 2xlw – AxCyt (mutant) + NO – Achromobacter xylosoxidans
1cgn, 1cgo - AxCyt
2xm0, 2xm4, 2xl8, 2xlh - AxCyt (mutant)
2xlm - AxCyt + NO
2j9b, 2j8w – Cyt – Rubrivivax gelatinosus
1gqa – RsCyt
1mqv, 1a7v – RpCyt – Rhodopseudomonas palustris
1eky – RcCyt]] - Rhodobacter capsulatus – NMR
1cpr, 1cpq, 1rcp – RcCyt
1nbb – RcCyt + cyanide
1e83, 1e84, 1e85, 1e86 – Cyt - Alcaligenes xylosoxidans
1jaf – Cyt – Rhodocyclus gelatinosus
1bbh – Cyt – Allochromatium vinosum
Cytochrome C’’Cytochrome C’’
1oae, 1gu2 – MmCyt – Methylophilus methylotrophus
1e8e – MmCyt - NMR
Cytochrome C1Cytochrome C1
3cx5, 3cxh – yCyt in complex III
2ibz - yCyt in complex III + inhibitor
1kyo - yCyt in Bc1 complex
1kb9 – yCyt in Bc1 complex residues 17-368
1ezv - yCyt in Bc1 complex + antibody FV fragment
3h1h, 1bcc - cCyt in Bc1 complex – chicken
3h1i, 2bcc, 3bcc - cCyt in Bc1 complex + inhibitor
2qjk, 2qjp, 2qjy – RsCyt in Bc1 complex + inhibitor
2fyn - RsCyt in Bc1 complex (mutant)
1l0n, 1be3, 1bgy, 1qcr – bCyt in Bc1 complex
2fyu - bCyt in Bc1 complex (mutant) + inhibitor
1sqp, 1sqq, 1sqv, 1sqx, 2a06, 1sqb, 1pp9, 1ppj, 1ntk, 1ntm, 1p84, 1l0l - bCyt in Bc1 complex + inhibitor
1ntz, 1nu1 - bCyt in Bc1 complex + substrate
1zrt - RcCyt in Bc1 complex + inhibitor
Cytochrome C2Cytochrome C2
1c2r - RcCyt
1vyd – RcCyt (mutant)
1c2n – RcCyt - NMR
1l9b, 1l9j – RsCyt in photosynthetic reaction center
2cxb, 1cxc, 1cxa - RsCyt
1jdl – Cyt – Rhodospirillum centenum
2c2c, 3c2c – Cyt – Rhodospirillum rubrum
1i8o, 1hh7 – RpCyt
1hro – Cyt – Rhodopila globiformis
1cot – PdCyt - Paracoccus denitrificans
1cry - RvCyt
Cytochrome C3Cytochrome C3
2ksu, 1up9, 1upd, 1gmb, 1gm4, 1i77, 3cyr – DdCyt
2kmy – DdCyt – NMR
2k3v – Cyt – Shewanella frigidimarina
1m1p, 1m1r - Cyt tetraheme – Shewanella oneidensis
1j0o, 2cth, 2cdv - DvCyt tetraheme
2z47, 2yyw, 2yyx, 2yxc, 2ffn, 2ewi, 2ewk, 2ewu, 1wr5, 1j0p, 1mdv, 2cym – DvCyt tetraheme (mutant)
1gx7 – DvCyt + hydrogenase
1gyo, 1wad - Cyt di-tetraheme – Desulfovibrio gigas
2bq4, 3cao, 3car – Cyt – Desulfovibrio africanus
1w7o - Cyt – Desulfomicrobium baculatus
1aqe – DnCyt (mutant) – Desulfomicrobium norvegicum
1czj, 2cy3 - DnCyt
Cytochrome C4Cytochrome C4
1m6z, 1m70, 1etp – PsCyt – Pseudomonas stutzeri
1h1o – Cyt - Acidithiobacillus ferrooxidans
Cytochrome C5Cytochrome C5
1cc5 – Cyt – Azotobacter vinelandii
Cytochrome C6Cytochrome C6
3ph2 – Cyt (mutant) – Phormidium laminosum
3dr0 – SyCyt – Synechococcus
3dmi – Cyt – Phaeodactylum tricornutum
2zbo – Cyt – Hizikia fusiformis
2v07, 2dge – AtCyt residues 71-175 – Arabidopsis thaliana
2ce0, 2ce1 - AtCyt residues 71-175 (mutant)
2v08 – Cyt – Phormidium laminosum
1ls9 – Cyt – Cladophora glomerata
1kib, 1f1f – AmCyt – Arthrospira maxima
1gdv – Cyt – Porphyra yezoensis
1a2s, 1ced – MbCyt – Monoraphidium braunii – NMR
1ctj - MbCyt
1c6s – Cyt – Cyanobacterium synechococcus]] - NMR
1c6o, 1c6r – Cyt – Scenedesmus obliquus
Cytochrome C7Cytochrome C7
3h33, 3h34, 3h4n, 3bxu – Cyt – Geobacter sulfurreducens
1lm2, 1l3o, 1kwj, 1f22, 1ehj – DaCyt – Deulfurmonas acetoxidans – NMR
1hh5 - DaCyt
Cytochrome C549Cytochrome C549
Cytochrome C550Cytochrome C550
3arc, 3prq, 3prr, 3kzi, 3a0b, 3a0h, 3bz1, 3bz2, 1izl – Cyt in photosystem II – Thermosynechococcus vulcanus
2axt, 1w5c, 1s5l - TeCyt in photosystem II – Thermosynechococcus elongatus
2bgv – PvCyt – Paracoccus versutus
2bh4, 2bh5 – PvCyt (mutant)
1mz4 – TeCyt
155c - PdCyt
Cytochrome C551Cytochrome C551
2zon – AxCyt + nitrite reductase
2gc7, 2gc4, 2mta – PdCyt + methylamine dehydrogenase + amicyanin
1cch, 1cor – PsCyt - NMR
1gks – Cyt – Ectothiorhodospira halophila - NMR
1new – DaCyt triheme]- NMR
2exv – PaCyt (mutant) – Pseudomonas aeruginosa
351c, 451c - PaCyt
2pac – PaCyt]] - NMR
1dvv - PaCyt (mutant) – NMR
Cytochrome C552Cytochrome C552
3l1t – EcCyt + sulfite – Escherichia coli
2rdz, 1gu6 – EcCyt
2rf7 – EcCyt (mutant)
3m97 – PdCyt soluble domain
3bnf - WsCyt + sulfite
3bng - WsCyt (mutant)
3bnh - WsCyt (mutant) + NO2
2e80 - WsCyt + NO2
2e81 - WsCyt + intermediate
3bnj - WsCyt (mutant) + sulfite
2ai5 – HtCyt – Hydrogenophilus thermophilus – NMR
2d0s, 1ynr – HtCyt
1ayg – Cyt - Hydrogenobacter thermoluteolus – NMR
1i6d, 1i6e, 1c7m – PdCyt functional domain – NMR
1ql3, 1ql4 - PdCyt functional domain
1dt1, 1qyz, 1r0q – TtCyt – Thermus thermophilus
2fwl – TtCyt + Cyt oxidase subunit II
1cno – Cyt – Pseudomonas nautica
Cytochrome C553Cytochrome C553
1b7v, 1c75 – BpCyt - Bacillus pasteuri
1k3h – BpCyt – NMR
1e08 – DdCyt + hydrogenase]- NMR
1n9c – Cyt – Sporosarcina pasteurii
1c53 - DvCyt
2dvh - DvCyt (mutant) - NMR
1dwl – DvCyt + ferredoxin I – NMR
1cyi, 1cyj – Cyt – Chlamydomonas reinhardtii
Cytochrome C554Cytochrome C554
2zzs – Cyt – Vibrio parahaemolyticus
1ft5, 1ft6, 1bvb – Cyt – Nitrosomonas europaea
Cytochrome C555Cytochrome C555
2zxy – Cyt – Aquifex aeolicus
2w9k – Cyt – Crithidia fasciculate
Cytochrome C556Cytochrome C556
1s05 – RpCyt - NMR
Cytochrome C558Cytochrome C558
2x5u, 2x5v – BvCyt in photosynthetic reaction center – Blastochloris viridis – Laue
2wjm, 2wjn, 3g7f, 3d38, 2jbl, 2i5n, 1vrn, 1r2c - BvCyt in photosynthetic reaction center
Cytochrome C NAPBCytochrome C NAPB
3ml1, 3o5a – Cyt + nitrate reductase catalytic subunit – Ralstonia eutropha
Cytochrome CLCytochrome CL
2d0w – Cyt – Hyphomicrobium denitrificans
2c8s – MeCyt – Methylobacterium extorquens
Cytochrome CC3Cytochrome CC3
Cytochrome CD1Cytochrome CD1
1gq1, 1h9x, 1h9y, 1hcm, 1qks – Cyt – Paracoccus pantotrophus
1gjq – PaCyt
1dy7 – PaCyt + CO
1e2r – PdCyt + CN
Cytochrome CHCytochrome CH
1qn2 – MeCyt
ReferencesReferences
- ↑ Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001 Nov 2;313(4):903-19. PMID:11697912 doi:10.1006/jmbi.2001.5080
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 Stelter M, Melo AM, Pereira MM, Gomes CM, Hreggvidsson GO, Hjorleifsdottir S, Saraiva LM, Teixeira M, Archer M. A Novel Type of Monoheme Cytochrome c: Biochemical and Structural Characterization at 1.23 A Resolution of Rhodothermus marinus Cytochrome c. Biochemistry. 2008 Oct 15. PMID:18855424 doi:10.1021/bi800999g
- ↑ 3.0 3.1 3.2 Reedy CJ, Gibney BR. Heme protein assemblies. Chem Rev. 2004 Feb;104(2):617-49. PMID:14871137 doi:10.1021/cr0206115
- ↑ 4.0 4.1 4.2 Ambler RP. Sequence variability in bacterial cytochromes c. Biochim Biophys Acta. 1991 May 23;1058(1):42-7. PMID:1646017
- ↑ Cookson DJ, Moore GR, Pitt RC, Williams RJP, Campbell ID, Ambler RP, Bruschi M, Le Gall J. Structural homology of cytochromes c. Eur J Biochem. 1978 Feb;83(1):261-75.
- ↑ Than ME, Hof P, Huber R, Bourenkov GP, Bartunik HD, Buse G, Soulimane T. Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing. J Mol Biol. 1997 Aug 29;271(4):629-44. PMID:9281430 doi:10.1006/jmbi.1997.1181
- ↑ Soares CM, Baptista AM, Pereira MM, Teixeira M. Investigation of protonatable residues in Rhodothermus marinus caa3 haem-copper oxygen reductase: comparison with Paracoccus denitrificans aa3 haem-copper oxygen reductase. J Biol Inorg Chem. 2004 Mar;9(2):124-34. Epub 2003 Dec 23. PMID:14691678 doi:10.1007/s00775-003-0509-9
- ↑ Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 2001 Jun 1;1505(2-3):185-208. PMID:11334784
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 Karp, Gerald (2008). Cell and Molecular Biology (5th edition). Hoboken, NJ: John Wiley & Sons. ISBN 978-0470042175.