Fumarase 2
This is a placeholderThis is a placeholder
This is a placeholder text to help you get started in placing a Jmol applet on your page. At any time, click "Show Preview" at the bottom of this page to see how it goes.
Replace the PDB id (use lowercase!) after the STRUCTURE_ and after PDB= to load and display another structure.
| |||||||||
| 3cin, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | TM1419, TM_1419 (Thermotoga maritima MSB8) | ||||||||
| Activity: | Inositol-3-phosphate synthase, with EC number 5.5.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
FumaraseFumarase
OverviewOverview
Fumarase, also known as fumarate hydratase, functions as an enzyme in the metabolic pathway known as the Kreb’s cycle, or citric acid cycle. As the seventh step in the pathway, fumarase catalyzes the reversible reaction converting fumarate to S-malate. It metabolizes Fumarate in the cytosol, which becomes a byproduct of the urea cycle and amino acid catabolism. It catalyzes the addition of water to make S-Malate; therefore, the mechanism of fumarase in the reaction involves hydration of fumarate in order to form S-malate.
Stucture and ClassificationStucture and Classification
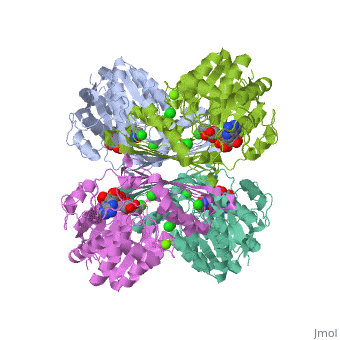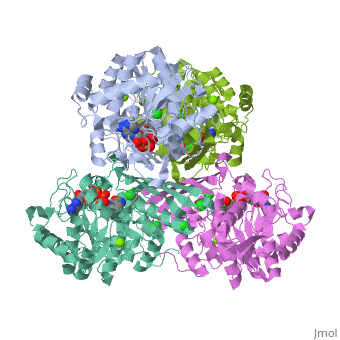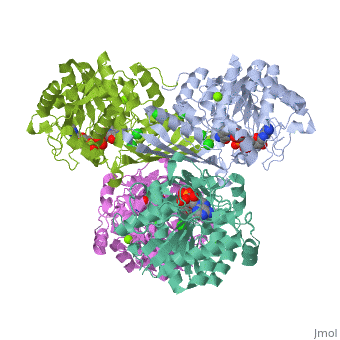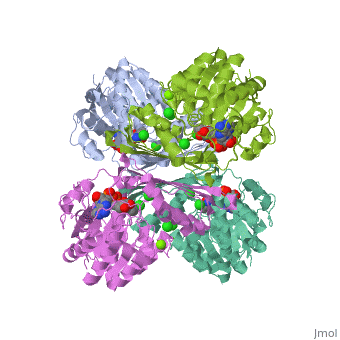
Fumarase is classified as an all alpha protein which belongs to the L-aspartase/fumarase family, and the enzyme specifically consists of four identical subunits which form a tetramer. From the four subunits, fumarase has three domains which comprise two binding sites: the active site and B site. Although the active site has a mostly solid structure and shifts very little when it binds, the B site shifts substantially more upon binding, and this shift helps regulate affinity for molecule binding at the active site (Weaver 2005).
Mechanism of ReactionMechanism of Reaction
Fumarase has the ability to catalyze the hydration of fumarate to malate or the dehydration of malate to fumarate. The mechanism of fumarase in the hydration and dehydration reaction pathways remains simple, only involving three steps. In the dehydration reaction, fumarase deprotonates a carbon atom on malate to form a carbanion (Beechmans 1998). This deprotonation results in an aci-carboxylate intermediate (Weaver 2005). After the intermediate forms, the acidic proton from the initial step removes the hydroxide group from the aci-carboxylate intermediate to form fumarase which then detaches from the active site of fumarase, completing the reaction (Rose, Weaver, 2004). The active site binds with the substrate via Asn326 and Lys324 residues which function in the binding process. The B site also can bind with S-malate, and the residues His129, Asn131, Asp132, and Arg126 contribute to binding (Weaver 2005). The B site is located in a π-helix turn between the active site and solvent. Two hydrogen bonds initiate the binding of Asn131 and Asp132 residues with S-malate (Rose, Weaver, 2004).
Enzyme KineticsEnzyme Kinetics
Fumarase catalyzes the reversible reaction between S-malate and fumarate in the citric acid cycle for cellular metabolism. When it catalyzes the addition of water to S-malate in order to form fumarate, the Km and Vmax values are 0.30 mM and 129 s^-1, respectively. The reverse reaction (dehydration of fumarate to form S-malate) has a Km of 0.10 mM and Vmax of 60 s^-1 (Rose, Weaver 2004). Thus, the forward pathway (fumarate to S-malate) is favored because of its higher Km and Vmax values. According to Beechmans (1998), fumarase kinetics normally follows Michaelis-Menten kinetic plots with low concentrations of substrate, but high concentrations influence the enzyme’s activity. Enzyme kinetics studies with fumarase mutants differ from wild type fumarase kinetics when the mutations alter amino acid residues involved in binding at the active site and B site. Also, the Michaelis-Menten kinetics plots for fumarase mutants exhibit a sigmoidal curve which suggests the presence of cooperativity in the enzymes activity.
Name of enzyme: Fumarase or Fumarate hydratase
Pathway and reaction catalyzed with metabolite structures: Fumarase is used in the citric acid cycle to conduct a transition step in the production of energy to make NADH. This is a reversible reaction. Conversion of Fumarate to S-Malate using Fumarase: Image:400px-Reaction1.png
Other interesting information: Fumarase is dominant in fetal and adult tissues and largely expressed in the skin, parathyroid, lymph, and colon There are two classes of Fumarases, which depend on the arrangement of their relative subunit, their metal requirement, and their thermal stability. Class I Fumarases can change their state or become inactive when exposed to heat or radiation. They are sensitive to superoxide anions and Fe2+ dependent. Class II Fumarases are found in eukaryotes and prokaryotes. They are iron-independent and thermal-stable. Fumarase deficiency is an autosomal recessive metabolic disorder distinguished by a deficiency of the enzyme Fumarate hydratase and indicated by an excess of Fumaric acid in the urine. It is common of infants with neurologic abnormalities and its potential causes include cytosolic and mitochondrial forms of Fumarase.