Lu sandbox 1
E1 Component of Multienzyme Pyruvate Dehydrogenase ComplexE1 Component of Multienzyme Pyruvate Dehydrogenase Complex
| |||||||||
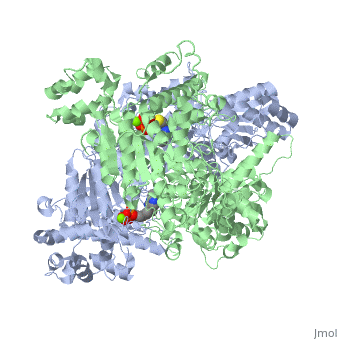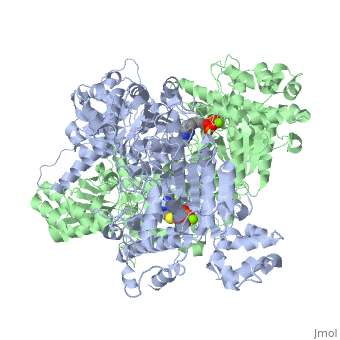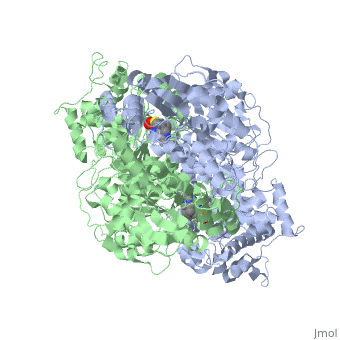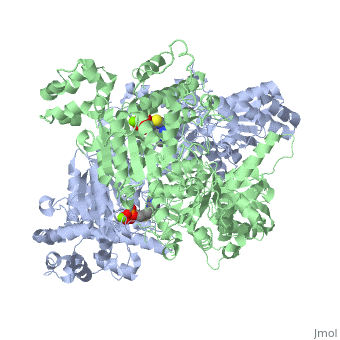
| 1l8a, resolution 1.85Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Pyruvate dehydrogenase (acetyl-transferring), with EC number 1.2.4.1 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Pyruvate dehydrogenase (E1) is one of three main components of the multienzyme complex pyruvate dehydrogenase. It accompanies dihydrolipoyl transacetylase (E2) and dihydrolipoyl dehydrogenase (E3) in comprising this multienzyme complex. Together, these enzymes are responsible for synthesizing acetyl-CoA from pyruvate just prior to entrance into the Krebs cycle.
StructureStructure
The E. coli enzyme complex has a weight of approximately 4600-kD and a diameter of about 300 angstroms. 24 of the 60 subunits within the complex are E1 proteins. Pyruvate dehydrogenase (E1) falls within the class of alpha and beta proteins[1], containing , and its structure in E. coli has been solved to a resolution of 1.85 angstroms. That study found that E1 is a homodimer with a molecular weight of 99474 containing α/β folds and bearing two catalytic sites located at the interface between subunits. Each polypeptide chain of E1 consists of 886 residues[2]. The structure shown is the E. coli E1 pyruvate dehydrogenase component, PDB code 1l8a[3].
Reactions and MecahanismReactions and Mecahanism
The multienzyme complex together catalyzes five distinct reactions in the conversion of pyruvate to acetyl-CoA. The overall result is described by the following reaction:
Pyruvate + CoA + NAD+ ==> Acetyl-CoA + CO2 + NADH
However, pyruvate dehydrogenase (E1) is responsible for only the first two of the five reactions. The first of these is the decarboxylation of pyruvate and coupling of thiamine pyrophosphate (TPP) to form hydroxyethyl-TPP.
Pyruvate + TPP ==> Hydroxyethyl-TPP + CO2
The enzyme requires 2+ as cofactors for catalysis.

In this reaction, the ylide form of TPP attacks the electrophilic carbonyl group of pyruvate. This reflects the ability of TPP’s thiazolium ring, which primarily interacts with , to add to carbonyl groups. Decarboxylation of the resulting alkoxide yields an enol complex. This enol resonates to form the ylide form of hydroxyethyl-TPP[4]. During this reaction, the catalytic Mg2+ ion coordinates octahedrally with three protein ligands: , which bind TPP, and [2].
The second E1-catalyzed reaction is the transfer of the hydroxyethyl group to the lipoamide group of the next enzyme, dihydrolipoyltransacetylase (E2). The E2 lipoamide group consisting of lipoic acid linked to the amide group of a Lys residue. Lipoic acid contains a reactive cyclic disulfide that is reversibly reduced to give dihydrolipoamide. The ylide form of the hydroxyethyl group of the hydroxyethyl-TPP complex attacks this disulfide bond. TPP is then eliminated as it detaches with E1 and subsequently binds to the next pyruvate molecule[4].
E1-Hydroxyethyl-TPP + E2-Lipoamide ==> E1-TPP + Acetyl-dihydrolipoamide-E2
RegulationRegulation
Pyruvate dehydrogenase plays a key role in the regulation of the Krebs cycle. The reactions catalyzed by the pyruvate dehydrogenase complex constitute the only biological pathway for acetyl-CoA synthesis from pyruvate. It is thus crucial that these reactions be precisely controlled.
One method of regulation is product inhibition by NADH and acetyl-CoA. NADH is a product of reactions catalyzed by dihydrolipoyl dehydrogenase (E3), while acetyl CoA is a product of the aforementioned dihydrolipoyl transacetylase (E2). Both compounds compete for active sites on their respective enzymes. NADH competes with NAD+ for E3 active site, while acetyl-CoA competes with CoA for E2 active site. High NADH levels keep E3 in its reduced form, thus the E2 lipoamide group stays in its reduced state. This in turn prevents E1 from transferring the hydroxyethyl group to E2. Consequently, E1 activity is reduced. Similarly, Acetyl CoA reduces E1 activity by occupying binding sites so less pyruvate binds to E1. Thus, high relative NADH and Acetyl-CoA concentrations regulate E1 activity through product inhibition.
These two compounds also activate the pyruvate dehydrogenase kinase associated with the enzyme complex[4]. This results in phosphorylation of three different E1 serine residues (Ser 203, Ser 264, Ser 271) in human E1 and enzyme inactivation[5]. Enzyme regulation through phosphorylation by pyruvate dehydrogenase kinase and dephosphorylation pyruvate dehydrogenase phosphatase has been implicated as a target for treating cancer, heart ischemia, and diabetes[6].
KineticsKinetics
E. coli pyruvate dehydrogenase binding constant and maximum velocity values have been reported as Km = 0.3 mM and Vmax = 5,500 kat/mol (37 degrees C, pH = 7.6, 5 microM pyruvate, and 3 mg/L protein). The multienzyme complex exhibits postive cooperative binding (Hill constant = 1.9)[7].
ReferencesReferences
- ↑ Protein: Pyruvate dehydrogenase E1-beta, PdhB, C-terminal domain from Bacillus stearothermophilus. (2009). Retrieved from http://scop.mrc-lmb.cam.ac.uk
- ↑ 2.0 2.1 Arjunan P, Nemeria N, Brunskill A, Chandrasekhar K, Sax M, Yan Y, Jordan F, Guest JR, Furey W. Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 A resolution. Biochemistry. 2002 Apr 23;41(16):5213-21. PMID:11955070
- ↑ Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
- ↑ 4.0 4.1 4.2 Voet, D., Voet, J.G., and Pratt, C.W. (2008). Fundamentals of biochemistry. Hoboken, NJ: John Wiley and Sons, Inc.
- ↑ Korotchkina LG, Patel MS. Probing the mechanism of inactivation of human pyruvate dehydrogenase by phosphorylation of three sites. J Biol Chem. 2001 Feb 23;276(8):5731-8. Epub 2000 Nov 22. PMID:11092882 doi:10.1074/jbc.M007558200
- ↑ Roche TE, Hiromasa Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell Mol Life Sci. 2007 Apr;64(7-8):830-49. PMID:17310282 doi:10.1007/s00018-007-6380-z
- ↑ Bisswanger, H. Substrate specificity of the pyruvate dehydrogenase complex from escherichia coli. J Biol Chem. 1981. Jan 25;256(2):815-822.
This is a placeholder text to help you get started in placing a Jmol applet on your page. At any time, click "Show Preview" at the bottom of this page to see how it goes.
Replace the PDB id (use lowercase!) after the STRUCTURE_ and after PDB= to load and display another structure.