Sandbox423
This sandbox is in use until August 1, 2011 for UMass Chemistry 423. Others please do not edit this page. Thanks!
L Thompson 3/16/11
Team & Protein selection, due 3/30/11
- Form teams of 4 people; include both chemistry and chemical engineering majors on each team. Start by finding a protein-drug complex with a known structure that interests your team (see assignment sheet instructions). Check the list below to see if another team has already chosen this protein. If not, start a new sandbox page (just try sandbox## in the search box to find an unused number) and add a link for your team/protein to our class list below (use editing button above (Ab) or follow my model).
- Copy the message at the top of this page into your sandbox page to "reserve" this sandbox for this course.
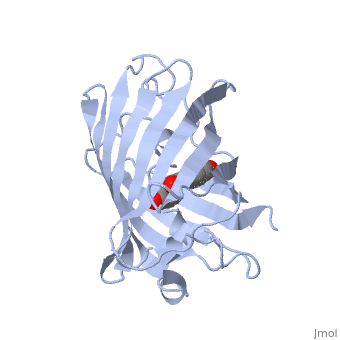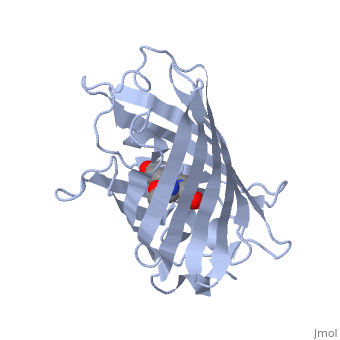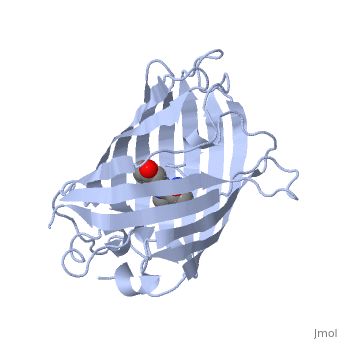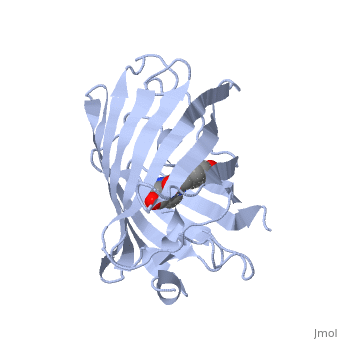
- Find the pdb id for your protein-drug complex in the Protein Data Bank. I looked for the earliest GFP structure I could find in the protein data bank and found 1ema, then followed the directions below to pull up a rotating GFP on this page. You can use the edit button on any page to find out how other users created effects that you see.
- Replace the PDB id (use lowercase!) after the STRUCTURE_ and after PDB= to load
and display another structure.
Project completion, due 4/22/11 Your proteopedia page should be organized into the following 4 required sections, with each team member responsible for one of these sections of the team project.
1. Introduction
- Introduce the protein function and the disease treated by the drug. This must be written in your own words with citations to your sources.You cannot include a copyrighted figure unless you request information to use it.
2. Overall structure
- Describe the overall structure of your protein in words and make "green scenes" to illustrate your points. What elements of secondary structure are present (ie 5 alpha helices and 2 beta strands) and how are they organized? Above illustrates the start of an "overall structure" section on GFP. Additional description and green scenes could illustrate the polar/nonpolar distrubution of amino acids (is the inside of the barrel polar or nonpolar?), packing of amphipathic elements, etc.
3. Drug binding site
- Describe features of the drug binding site in words and make "green scenes" to illustrate your points. What interactions stabilize binding of this molecule to the protein.
4. Additional features
- Describe and use green scenes to illustrate additional features of the protein. What you do here depends on what information is available. If a structure of the protein-substrate complex is available, you could compare protein interactions with the substrate vs. with the drug. If the drug is a transition state inhibitor, explain and illustrate that (include figures describing the reaction).
Credits -- at the end list who did which portion of the project:
- 1. Introduction -- name of team member
- 2. Overall structure -- name of team member
- 3. Drug binding site -- name of team member
- 4. Additional features -- name of team member
Overall StructureOverall Structure
| |||||||||
| 1ema, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | , | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
The of green fluorescent protein is an 11-stranded antiparallel beta barrel (yellow) surrounding a central helix (pink). The chromophore is part of the central helix. Information from 1ema.