Citrate Synthase

The Structure and Mechanism of Citrate SynthaseThe Structure and Mechanism of Citrate Synthase
|
Citrate synthase is an enzyme active in all examined cells, where it is most often responsible for catalyzing the first reaction of the citric acid cycle (Krebs Cycle or the tricarboxylic acid [TCA] cycle): the condensation of acetyl-CoA and oxaloacetate to form citrate. Although in eukaryotes it is a mitochondrial enzyme, and in fact, is often used as a enzyme marker for intact mitochondria, it is encoded by nuclear DNA[1]. The standard free energy change (ΔG°’) for the citrate synthase reaction is -31.5kJ/mol [2]. This negative free energy value means that citrate synthase is likely to function far from equilibrium under physiological conditions, and is thus a rate-determining enzyme in the citric acid cycle.
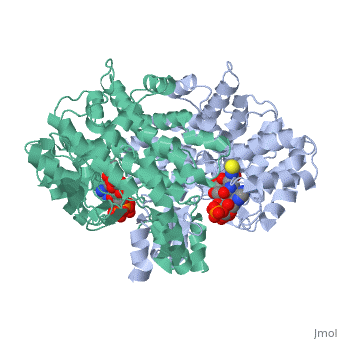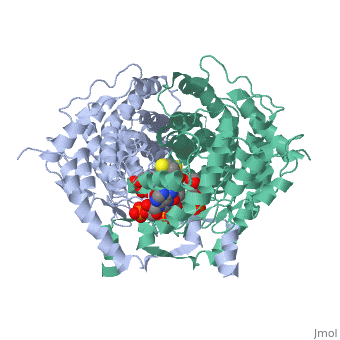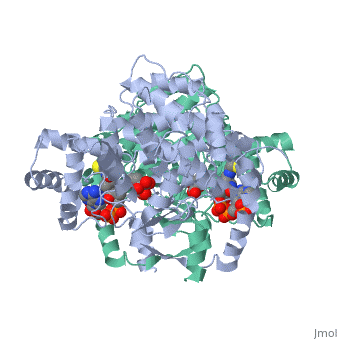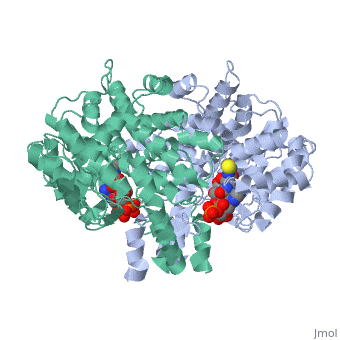
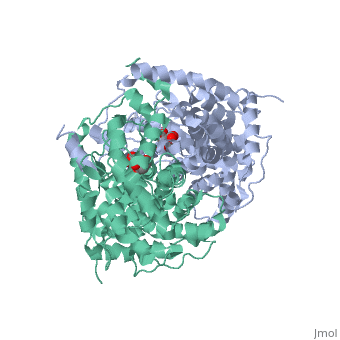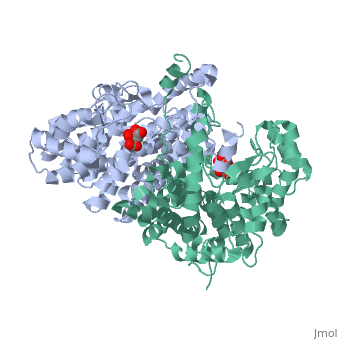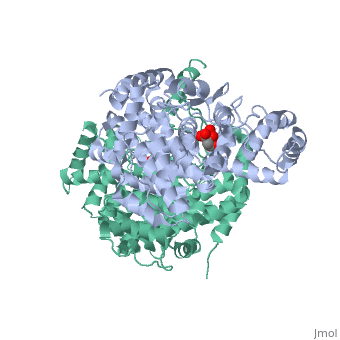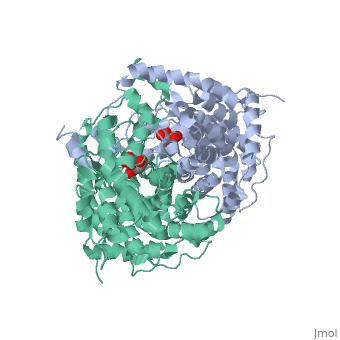
Structure: Biologically, citrate synthase exists as a
of a single amino acid chain . Each identical subunit consists of a large and a small domain, and is comprised almost entirely of α helices (making it an all α protein). In its free enzyme state, citrate synthase exists in , with its two domains forming a cleft containing the substrate (oxaloacetate) binding site (PDB: 1cts) [3][4]. When oxaloacetate binds, the smaller domain undergoes an 18° rotation, sealing the oxaloacetate binding site[5] and resulting in the (PDB: 2cts)[3]. The dramatic conformational change is best illustrated via , and be sure to view as well to get a full sense of the structural change. The conformational change not only prevents solvent from reaching the bound substrate, but also generates the acetyl-CoA binding site. This presence of “open” and “closed” forms results in citrate synthase having Ordered Sequential kinetic behavior [2].
| |||||||||||||
 | |||||||||||||
|
|
Literature and NotesLiterature and Notes
- ↑ "Citrate Synthase -." Wikipedia, the Free Encyclopedia. Web. 22 Mar. 2010.
- ↑ 2.0 2.1 Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008.
- ↑ 3.0 3.1 3.2 Remington S, Wiegand G, Huber R. Crystallographic refinement and atomic models of two different forms of citrate synthase at 2.7 and 1.7 A resolution. J Mol Biol. 1982 Jun 15;158(1):111-52. PMID:7120407
- ↑ In this structure 1cts, citrate, the resulting product of the conversion, is actually bound where oxaloacetate binds.
- ↑ Bayer E, Bauer B, Eggerer H. Evidence from inhibitor studies for conformational changes of citrate synthase. Eur J Biochem. 1981 Nov;120(1):155-60. PMID:7308213
- ↑ Karpusas M, Branchaud B, Remington SJ. Proposed mechanism for the condensation reaction of citrate synthase: 1.9-A structure of the ternary complex with oxaloacetate and carboxymethyl coenzyme A. Biochemistry. 1990 Mar 6;29(9):2213-9. PMID:2337600
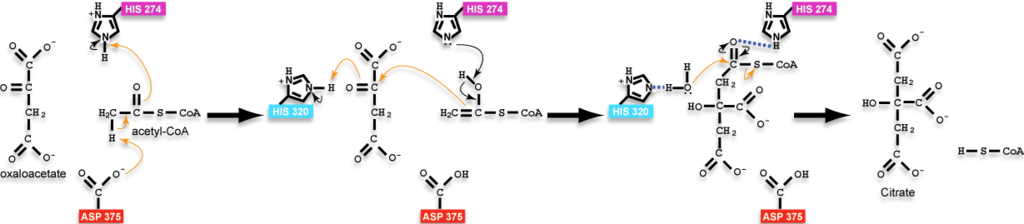
- ↑ 5cts as the state preceding condensation with oxaloacetate and a non-reactive version of acetyl-CoA bound, 6cts as the state containing the bound intermediate, and 3cts as the complex with the products. Positions of hydrogens on the ligands were calculated and added back to structures in the reaction scheme for instructional purposes and are not present in the experimentally-determined structures; additionally, arrows are drawn with atoms of the analog of acetyl-CoA to approximate the position of the reactive groups only as the reactive groups are not actually part of the analog or the molecules would have reacted; please, see the reaction scheme on this page for a more thorough accounting of the chemistry.
- ↑ Wiegand G, Remington SJ. Citrate synthase: structure, control, and mechanism. Annu Rev Biophys Biophys Chem. 1986;15:97-117. PMID:3013232 doi:http://dx.doi.org/10.1146/annurev.bb.15.060186.000525
- ↑ Kim KS, Rosenkrantz MS, Guarente L. Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol Cell Biol. 1986 Jun;6(6):1936-42. PMID:3023912
- ↑ Lewin AS, Hines V, Small GM. Citrate synthase encoded by the CIT2 gene of Saccharomyces cerevisiae is peroxisomal. Mol Cell Biol. 1990 Apr;10(4):1399-405. PMID:2181273
- ↑ Lee YJ, Hoe KL, Maeng PJ. Yeast cells lacking the CIT1-encoded mitochondrial citrate synthase are hypersusceptible to heat- or aging-induced apoptosis. Mol Biol Cell. 2007 Sep;18(9):3556-67. Epub 2007 Jul 5. PMID:17615299 doi:10.1091/mbc.E07-02-0118
Details of Structures FeaturedDetails of Structures Featured
1cts is a 1 chain structure of sequence from Sus scrofa. Full crystallographic information is available from OCA.
2cts is a 1 chain structure of sequence from Sus scrofa. Full crystallographic information is available from OCA.
3cts is a 1 chain structure of sequence from Gallus gallus. Full crystallographic information is available from OCA.
5cts is a 1 chain structure of sequence from Gallus gallus. Full crystallographic information is available from OCA.
6cts is a 1 chain structure of sequence from Gallus gallus. Full crystallographic information is available from OCA.
Additional StructuresAdditional Structures
See AlsoSee Also
External ResourcesExternal Resources
- Citrate Synthase: September 2007 Molecule of the Month as part of the series of tutorials that are at the RCSB Protein Data Bank and written by David Goodsell
- An interactive schematic animation of Citrate synthase's reaction mechanism from Lehninger's Principles of Biochemistry
- Citrate Synthase at Wikipedia