Jmol/Visualizing large molecules
|
Inadequate Memory May Preclude DisplayInadequate Memory May Preclude Display
Some molecular models ("molecules") are so large that they will not fit within the default amount of computer memory allocated to Jmol (which is the default amount of memory allocated to java). While it is possible to increase the memory allocated to java, most users will not do this, and hence, will not be able to display, in Proteopedia or Jmol, molecules that exceed a certain size.
SolutionsSolutions
Below are explained various strategies for reducing the sizes of large PDB files, enabling their main features to be displayed in the default Jmol/java memory. These strategies include displaying only the backbones (alpha carbons for proteins and phosphorus atoms for nucleic acids), and displaying one, or a subset, of the models in multiple-model files.
Maximum Size Per ModelMaximum Size Per Model
99,999 Atoms99,999 Atoms
Strictly speaking, the format of PDB files is limited to 99,999 atoms in a single model, because there are only 5 columns allocated to atom serial numbers. (Files in the mmCIF format can be read by Jmol, and do not suffer from this limitation.) 3cc2 is a model of a large ribosomal subunit containing 99,049 atoms (close to the limit for a single PDB file). Most likely it will display in Jmol when you go to that page. Jmol ignores the atom serial number, columns 7-11 in the PDB file, instead assigning its own atomIndex number, unique for each atom, and not redundant between models. Jmol can handle PDB files containing >100,000 atoms.
This limitation requires that models containing >=100,000 atoms be split into two or more PDB files, or else represented as artificially separated models in a single PDB file. These work-arounds are awkward for visualization. An example is the combination of portions of the two files 1jgo and 1giy for visualization of a complete Ribosome.
Rat Liver Vault, alpha carbon atoms only. | |
241,956 Atoms: Rat Liver Vault241,956 Atoms: Rat Liver Vault
The rat liver vault needed to be split into 3 PDB files: 2zuo, 2zv4, and 2zv5. Each file contains 80,652 atoms in 13 chains, for a total in the asymmetric unit of 241,956 atoms in 39 chains (A-Z, a-m). The biological unit contains 2 asymmetric units. Fortunately, the authors provide PDB files containing complete asymmetric units. However, these are 18 megabyte files, and do not fit in Jmol/java default memory. The methods explained below will enable you to visualize an asymmetric unit as alpha carbon atoms only (31,668 atoms). First, run the Jmol application. Now try these commands (the load command will take about a full minute):
- load http://www.protein.osaka-u.ac.jp/olabb/tsukihara/mvp/mvp_39mer.pdb filter "*.ca"
- color chain
62 Chains62 Chains
In the most recent update of the PDB data format specification (Version 3.2, October 2008), chain IDs (names) must be single alphanumeric characters (A-Z, a-z, 0-9). This permits a maximum of 62 chains. This limit is not much of a problem for asymmetric units. In January, 2011, there is only one PDB entry with 62 chains (2zkr), and 4 more with 55-60 chains.
Jmol can automatically apply a distinct color to each chain, up to 36 chains (Jmol Colors). However, it can distinguish 62 chains by selection (see set chainCaseSensitive).
Multiple Model FilesMultiple Model Files
The largest PDB files in the Protein Data Bank are those containing multiple models of large molecules. Since the atom serial numbers start at 1 in each model, these files can get very large (>1,000,000 atoms is possible). An example is 3ezb, which contains 40 models (determined by solution NMR). Each model contains 5,323 atoms (including 2,694 hydrogen atoms); the 40 model file contains 212,920 atoms, and the PDB file is 16.5 megabytes in size. When you visit the page 3ezb, the ensemble will fail to display, producing an "out of memory" error (unless you have allocated more than the default amount of memory to java on your computer). There are files in the PDB several-fold larger than 3ezb. For example, 2hyn is a 64 megabyte file containing 826,896 atoms in 184 models.
Displaying Only The First ModelDisplaying Only The First Model
Jmol can be instructed to load only the first model of a multiple-model PDB file. This is best done with the Jmol application (outside of Proteopedia). Later, the single model could be uploaded to Proteopedia for use in a scene.
- Run the Jmol Application in a working folder. Here are instructions.
- Demonstrate Out Of Memory: Type the following command into the white console window:
- load =2hyn
- The equal sign tells Jmol to obtain the PDB file from the Protein Data Bank. A red "OutOfMemory" error message should appear in Jmol in less than 30 seconds (depending on the speed of you Internet connection).
- Load The First Model: Type the following two commands into the white console window:
- zap
- load models {1 1 1} =2hyn
- In less than 30 seconds, the first model from the ensemble in 2hyn should appear in Jmol.
- Save The First Model: Type this command:
- write pdb 2hyn_model1.pdb
- Now you should find a new file 2hyn_model1.pdb in your working folder. You can load it with this command:
- load 2hyn_model1.pdb
- You can also upload it to Proteopedia for use in molecular scenes generated with Proteopedia's SAT.
Displaying Only Alpha Carbon AtomsDisplaying Only Alpha Carbon Atoms
With large multiple-chain assemblies, or multiple-model ensembles, typically you want to see only the backbone traces. Backbone traces can be visualized from only the alpha carbon atoms (or for nucleic acids, the phosphorus atoms). Jmol can extract ("filter") specified atoms from the PDB file, thereby saving memory. For example, 2hyn contains 4,494 atoms/model (half of which are hydrogen atoms), and 184 models, totalling 826,896 atoms. There are 260 alpha carbon atoms/model, or a total of 47,840 atoms. The alpha carbons represent less than 6% of the original atoms, or a nearly 20-fold reduction in memory requirements.
Using the Jmol application from your working folder (see instructions), enter this command:
- load =2hyn filter "*.ca"
"*.ca" means "all carbon alpha atoms". After about a full minute (depending on the speed of your Internet connection), a backbone trace of the first model will appear, which means that loading and filtering are complete. These commands will display the backbone traces for all 184 models:
- frame all
- color chain
If you wish, you can save the alpha-carbon atom models:
- write pdb 2hyn_ca_only.pdb
This file could be uploaded to Proteopedia for use in the SAT.
If your molecule contains nucleic acid, you will also want the nucleic backbone traces. The command (for PDB code 2o5i, 52,717 atoms including protein, DNA and RNA) is
- load =2o5i filter "*.ca, *.p"
Displaying Alpha Carbons For A Subset Of ModelsDisplaying Alpha Carbons For A Subset Of Models
Suppose that you want the alpha carbons for a subset of models in the published ensemble. You can get 16 models from the 184 models in 2hyn by taking either the first 16
- load models {1 16 1} =2hyn filter "*.ca"
of by taking every 12th model plus the last model
- load models {1 184 12} =2hyn filter "*.ca"
Biological AssembliesBiological Assemblies
The functional forms of molecules, often called biological units or biological assemblies, may contain many copies of the chains present in the published PDB file (the asymmetric unit).
Virus CapsidsVirus Capsids
An extreme example is a virus capsid. The capsid of the Simian Virus 40 (SV40) contains 360 copies of the VP1 protein chain, present in 6 copies in the published PDB file 1sva. An extremely simplified model of the capsid is displayed at SV40_Capsid_Simplified, but this model is oversimplified for some purposes, and required special techniques to construct. We can get the full capsid model from any of several servers. We recommend getting it from the ViperDB at Scripps, a server specialized in virus capsid structures.
The full SV40 capsid model (360 copies of the VP1 protein chain, minus hydrogen atoms) is a PDB file of 70 megabytes, much too large for default java/Jmol memory. Below are instructions for getting the much smaller alpha carbon atom model. These instructions should work for most virus capsids.
SV40 Capsid Alpha CarbonsSV40 Capsid Alpha Carbons
- Run the Jmol Application in a working folder. Here are instructions.
- Get the Address of the Capsid Structure: Go to ViperDB, and submit the PDB code 1sva. Right click on the link full capsid, then copy link location. For 1sva, it is http://viperdb.scripps.edu/OLIGOMERS/1sva_full.vdb.gz.
- Display The Alpha Carbons: In the Jmol application, enter these commands
- load http://viperdb.scripps.edu/OLIGOMERS/1sva_full.vdb.gz filter "*.ca"
- color chain
It may take about a minute for the first command to work. After the capsid appears, enter the second command -- it may take half a minute. The resulting display will include 123,420 atoms, which is about the maximum that Jmol can display in the default java memory.
If the SV40 full capsid display fails, try the half-capsid model (also available from a link at ViperDB). You will probably want to look at the half capsid anyway, as that shows the inside better than using Jmol's slab command.
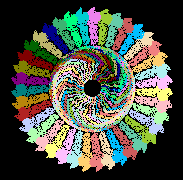
Human Heptatitis B Capsid Alpha CarbonsHuman Heptatitis B Capsid Alpha Carbons
A smaller virus capsid is human hepatitis B, 2g33. Its half-capsid (17,460 atoms) is displayed near the top of this page.
Non-Capsid Biological AssembliesNon-Capsid Biological Assemblies
This section is incomplete and remains under construction. Eric Martz 00:52, 3 January 2011 (IST)