Factor VIIa
This is a placeholderThis is a placeholder
This is a placeholder text to help you get started in placing a Jmol applet on your page. At any time, click "Show Preview" at the bottom of this page to see how it goes.
Replace the PDB id (use lowercase!) after the STRUCTURE_ and after PDB= to load and display another structure.
| |||||||||
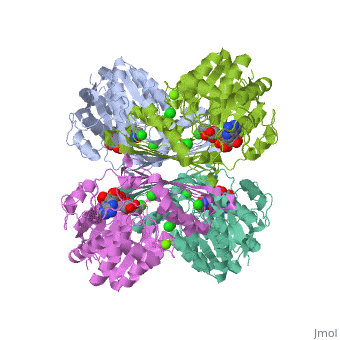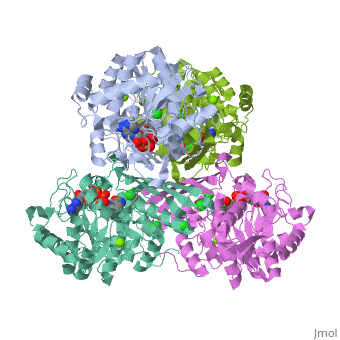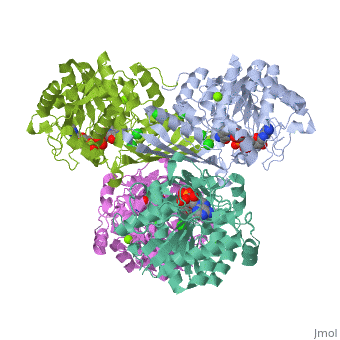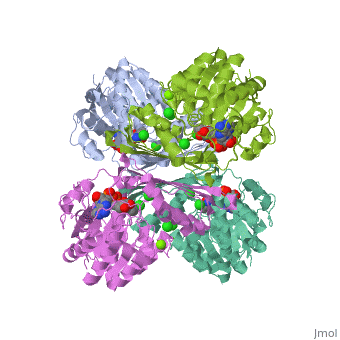
| 3cin, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | TM1419, TM_1419 (Thermotoga maritima MSB8) | ||||||||
| Activity: | Inositol-3-phosphate synthase, with EC number 5.5.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
FVIIaFVIIa
Factor VIIa (FVIIa) is a single chain trypsin-like serine protease (EC 3.4.21.21) of 406 residues. The FVII zymogen is a glycoprotein consisting of an amino-terminal (N-linked) γ-carboxyglutamic acid (Gla) domain followed by two epidermal growth factor-like (EGF) domains, a short linker peptide, and a carboxy terminal serine protease domain (Figure 1)[1]. The Gla domain is responsible for Ca++ binding leading to structural reorganization of FVIIa into a membrane binding protein2. The active form, FVIIa, is generated by a specific cleavage of a peptide bond between Arg-152 and Ile-153 at the end of the linker peptide by either factor Xa (FXa) or thrombin (IIa). This cleavage generates an N-terminal light chain of 152 residues linked to a heavy chain of 254 residues by a disulfide bridge3,4. Following cleavage the newly formed N-terminal inserts itself into a cavity, or the activation pocket, forming a salt bridge with Asp343 (Asp194 in trypsin).Formation of this salt bridge allows for the maturation of FVIIa to its active form. FVIIa alone is not very efficient in the insertion of the peptide, however, upon binding to TF the process becomes very efficient. The insertion of the N-terminus, therefore, facilitates the formation of both the S1 recognition pocket and the oxyanion hole. BACE1 (β-site of APP cleaving enzyme) also called β-Secretase and memapsin-2 is a 52 kD class I transmembrane aspartic acid protease that cleaves the Amyloid Precursor Protein (APP) in a rate limiting step that contributes to the accumulation of β-amyloid plaques in Alzheimer’s disease (AD). A subsequent cleavage by γ-secretase generates a 40 or 42 amino acid β-amyloid peptide. These peptides can form Aβ plaques that may have deleterious effects on neuronal function and contribute to pathologies of AD. Under normal conditions, BACE1 activity generates a monomeric and soluble Aβ peptide that may play a physiological role in decreasing excitotoxicity and neurotransmission at glutamatergic synapses. Additionally, α-secretase and γ-secretase cleave APP to generate p3 and the carboxy terminal fragment AICD in a non-amyloidogenic pathway. In AD, amyloidogenic pathways become preferential over non-amyloidogenic and Aβ plaques appear under increased levels of BACE1 catalytic activity.