100d
|
CRYSTAL STRUCTURE OF THE HIGHLY DISTORTED CHIMERIC DECAMER R(C)D(CGGCGCCG)R(G)-SPERMINE COMPLEX-SPERMINE BINDING TO PHOSPHATE ONLY AND MINOR GROOVE TERTIARY BASE-PAIRING
OverviewOverview
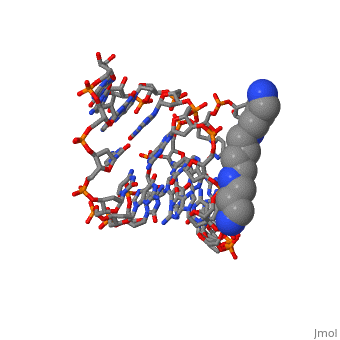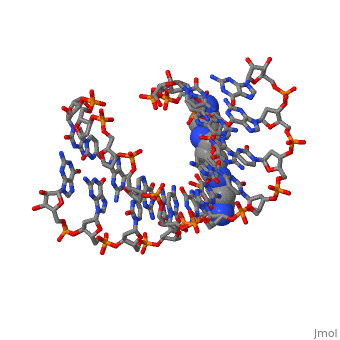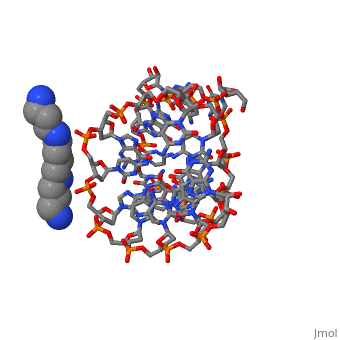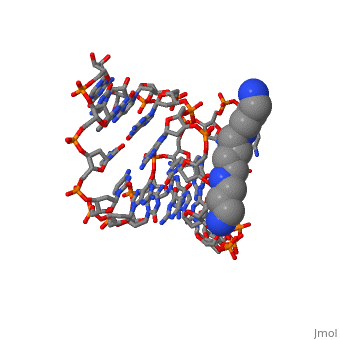
The crystal structure of the self-complementary chimeric decamer duplex, r(C)d(CGGCGCCG)r(G), with RNA base pairs at both termini, has been solved, at 1.9 A resolution by the molecular replacement method and refined to an, R value of 0.145 for 2,314 reflections. The C3'-endo sugar puckers of the, terminal riboses apparently drive the entire chimeric duplex into an A-DNA, conformation, in contrast to the B-DNA conformation adopted by the, all-deoxy decamer of the same sequence. Five symmetry related duplexes, encapsulate a spermine molecule which interacts with ten phosphate groups, both directly and through water molecules to form multiple ionic and, hydrogen bonding interactions. The spermine interaction severely bends the, duplexes by 31 degrees into the major groove at the fourth base ... [(full description)]
About this StructureAbout this Structure
100D is a [Protein complex] structure of sequences from [[1]] with SPM as [ligand]. Full crystallographic information is available from [OCA].
ReferenceReference
Crystal structure of the highly distorted chimeric decamer r(C)d(CGGCGCCG)r(G).spermine complex--spermine binding to phosphate only and minor groove tertiary base-pairing., Ban C, Ramakrishnan B, Sundaralingam M, Nucleic Acids Res. 1994 Dec 11;22(24):5466-76. PMID:7816639
Page seeded by OCA on Mon Oct 29 15:37:40 2007