Malate dehydrogenase
Malate DehydrogenaseMalate Dehydrogenase
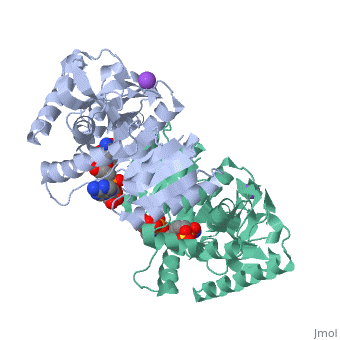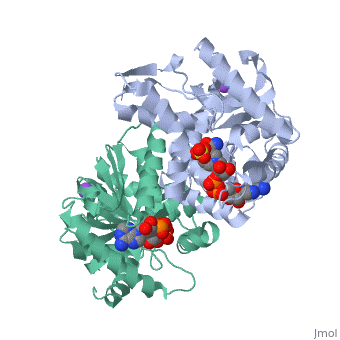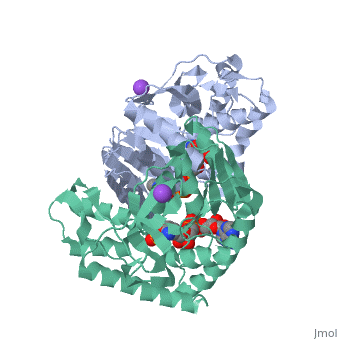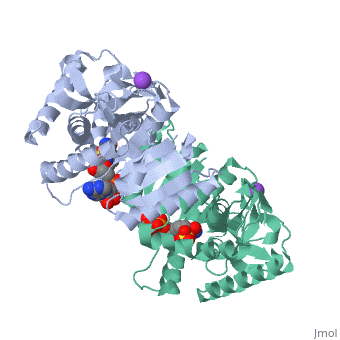
Malate Dehydrogenase (MDH)(PDB entry 2x0i) is most known for its role in the metabolic pathway in the tricarboxylic acid cycle[1] and is classified as a oxidoreductase[2]. The enzyme exists in two places inside a cell, in the mitochondria and cytoplasm. In the mitochondria, the enzyme catalyzes the reaction of malate to oxaloacetate; but in the cytoplasm, the enzyme catalyzes oxaloacetate to malate to allow transport. The enzyme malate dehydrogenase is composed of either a dimer or tetramer. During catalysis, the enzyme subunits are non-cooperative between active sites. The mitochondrial MDH is allosterically controlled bycitrate, but no other known metabolic regulation mechanisms have been discovered.
| |||||||
| 2x0i, resolution 2.91Å () | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||
| Activity: | Malate dehydrogenase, with EC number 1.1.1.37 | ||||||
| Related: | 2x06, 2x0j, 2x0n, 2x0r, 2x0s | ||||||
| |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
The secondary structure of a single subunit contains a 9 beta sheet parallel backbone wrapped by 9 large alpha helices. Near the sodium bound end, 4 small anti-parallel beta sheets and 1 small alpha helix enable a turn in the residue chain. Opposite the sodium bound ligand, 6 alpha helices point towards a common point, three on each side of the beta sheet backbone. The alpha helices form a small groove for a NAD+ cofactor to attach neat the beta sheeting. The structure most nearly resembles an alternating alpha/beta classification. As for the 3D structure, the enzyme forms a sort of crevice for the substrate to bind.
The mechanism of catalysis is dependent on several invariant residues. These residues are HIS 195 and ASP 168 which are involved in hydrogen bonding, ASP 53 associated with NAD+ binding, and a triad of arginine residues at 102, 109, and 171. During the conversation of malate to oxaloacetate, a key conformational change occurs on the binding of substrate in which a “loop” flips into an up position to block the active site from the solvent. When this occurs, the other residues in the active site are brought closer to the substrate to enable the conversion. R102 and R109 are involved in this loop flip and thus invariant. After the loop flip, the malate complex is stabilized via hydrogen bonding before accepting a proton transfer from HIS 195 to form oxaloacetate.
The evolutionary past of MDH shows a divergence to form lactate dehydrogenase (LDH) which functions in a very similar way to MDH. Although there is a very low sequence conservation among MDH and LDH’s the structure of the enzyme has remained relatively conserved. The key difference between the two is in the substate: LDH catalyzes pyruvate to lactate.