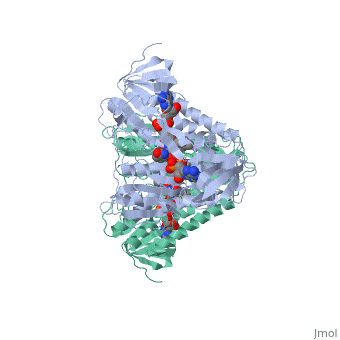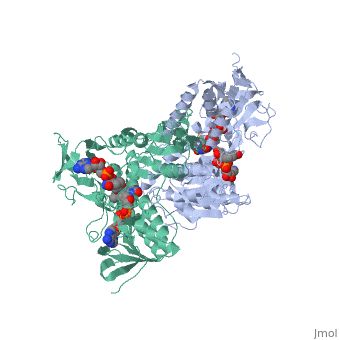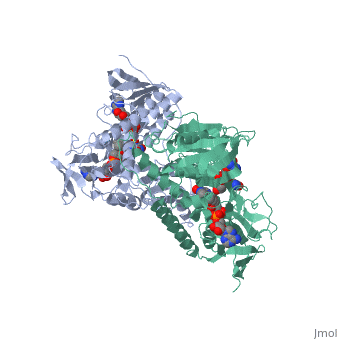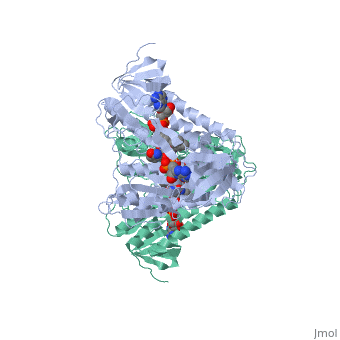
Evans sandbox 1
| |||||||||
| 1lvl, resolution 2.45Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Dihydrolipoyl dehydrogenase, with EC number 1.8.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
General Structure and InformationGeneral Structure and Information
Dihydrolipoamide dehydrogenase(E3), a component of the Saccharomyces cerevisiae and mammalian Pyruvate dehydrogenase complexes (PDC), anchors an E3 homodimer inside each of the 12 pentagonal faces of the 60-mer dihydrolipoamide acetyltransferase (E2)[1] PDC is the enzyme in the citric acid cycle responsible for the reaction converting Pyruvate to Acetyl CoA, NAD+ to NADH and H+ and the release of carbon dioxide. E3's cofactors include NAD+ and FAD, and unlike E1 and E2, there are equal subunits noncovalently bonded to the 60-meric transacetylase core in both prokaryotes and eukaryotes. The jmol image shown upon loading the page is the base subunit which is bonded to the core.
E3 is common to all a-ketoacid dehydrogenase complexes. Errors in the gene coding human E3 cause combined deficiencies in a-ketoacid dehydrogenase complexes manifested by lactic acidemias and Maple Syrup Urine Disease[2]. A subset of the human E3 mutations has been suggested to occur at the homodimer interface or at the putative E3/E3BP interaction surface
E3 is synthesized as a precursor form in the cytoplasm and imported into mitochondria, and the mature E3 migrates as a 55 kDa polypeptide in SDS-PAGE though its calculated molecular mass with one molecule of FAD is 51 kDa.
Structure in Pyruvate Dehydrogenase ComplexStructure in Pyruvate Dehydrogenase Complex
In E. coli this complex exists as 24 E2 proteins arranged in a cube, surrounded by 12 E1 proteins and 12 E3 proteins. Dihidrolipoamide dehydrogenase (E3) binds to the pyruvate dehydrogenase complex (and the center of the cube of E2 proteins) through a .‘[1]’ The E3 binding protein is a completely separate protein from E3, but serves to connect the E3 polypeptides to the overarching structure. The active site includes an FAD group, as well as forming a disulfide bond. When the substrate is not present, “covers” the catalytic site from being exposed to solvents. Dihidrolipoamide dehydrogenase (E3) is a SCOP alpha and beta (a/b) class protein of the FAD/NAD(P)-binding domain fold.
MechanismMechanism
The redox reaction occurs through the influence of , between which there is a disulfide bond within a distorted alpha helix. This redox active disulfide bond becomes reduced in order to reoxidize the E2 enzyme of the multienzyme complex.‘[2]’ E2 donates protons and electrons to E3 in order to complete its catalytic cycle. The E3 enzyme’s flavin ring (FAD) funnels electrons from the disulfide bond to itself, , reoxidizing the E3, regenerating FAD, producing NADH, and leaving it ready for the beginning of its catalytic cycle again.
A 16-amino acid leader sequence addition changes the kinetic mechanism of human E3 so that it resembles the mixed sequential and ping-pong mechanism of Glutathione Reductase‘[3]’
RegulationRegulation
The regulation of E3 kinetically comes through regulation of the entire Pyruvate Dehydrogenase complex. As would be expected, one of the main regulators is the presence of its product, acetyl-CoA as well as NADH. NADH competes with NAD+ with the binding site on E3, therefore regulating the entire pyruvate dehydrogenase complex when NADH is in high concentration.‘[4]’
- ↑ Brautigam CA, Wynn RM, Chuang JL, Machius M, Tomchick DR, Chuang DT. Structural insight into interactions between dihydrolipoamide dehydrogenase (E3) and E3 binding protein of human pyruvate dehydrogenase complex. Structure. 2006 Mar;14(3):611-21. Epub 2006 Jan 26. PMID:16442803 doi:10.1016/j.str.2006.01.001
- ↑ Voet, Donald et al. 2008. Fundamentals of Biochemistry. 3rd ed. pp.570-575
- ↑ Kim, Hakjung Expression of cDNA Sequences Encoding Mature and Precursor Forms of Human Dihydrolipoamide Dehydrogenase in Escherichia coli. Journal of Biological Chemistry. 266, 15. 1991.
- ↑ Voet, Donald et al. 2008. Fundamentals of Biochemistry. 3rd ed. p.585