Eric Comeau/ sandbox
Sickle Cell HemoglobinSickle Cell Hemoglobin
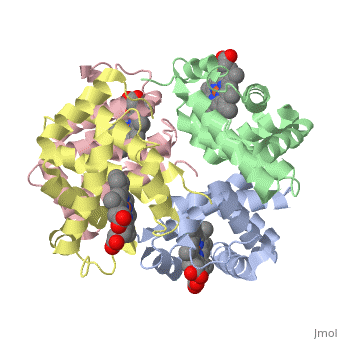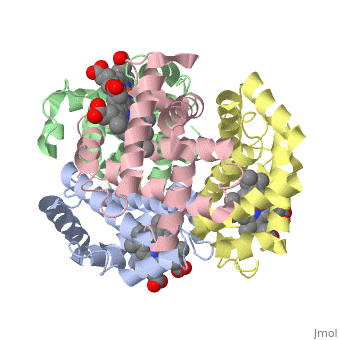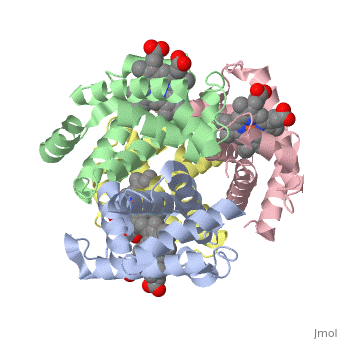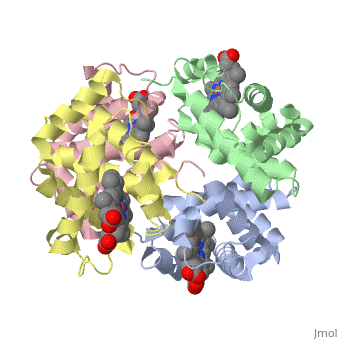
This is a default text for your page Eric Comeau/ sandbox. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue. Physical modelThe 3D-printed models are of the tetramer, with one beta subunit valine marked with a blue rod. One hydrophobic surface patch is marked with a hole. In the browser, the is shown in red (alpha subunits) and orangered (beta subunits), with both valines shown in blue. In a second scene, are shown. You can see that one of the four valines is buried in the contact area between the two tetramers. The (shown in gray) consists of Phe 85 and Leu 88 side chains in an indentation of the protein's surface. Sickle Cell Anemia
The is shown in gray, the mutations to valine in blue.
RelevanceStructural highlightsThis is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
|
| ||||||||||
ReferencesReferences
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644