Introduction to protein structure
Levels of Protein StructureLevels of Protein Structure
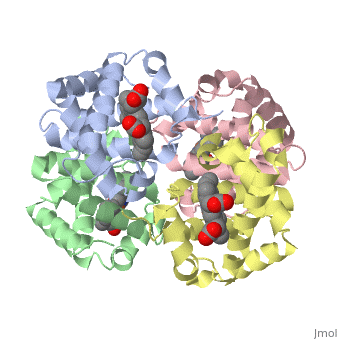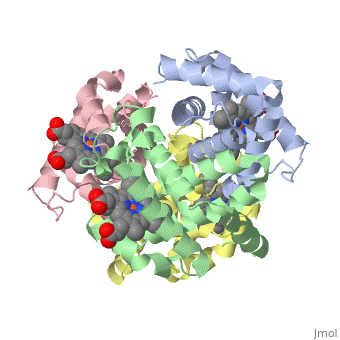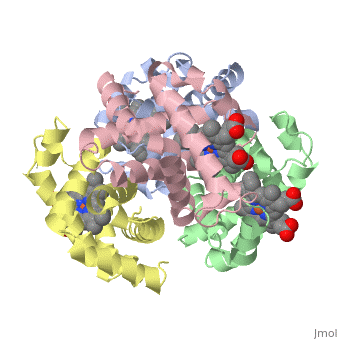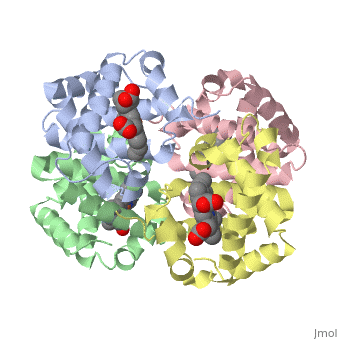
This tutorial illustrates some basic properties of protein structure and useful commands in Jmol and Proteopedia. Clicking the green links changes the view in the structure box to illustrate the principle described by the text. Proteins are condensation polymers of amino acids. The is the amino acid sequence. The is the local structure over short distances. This level of structure is stabilized by along the . These secondary structures to form the overall form of the entire peptide chain, called the . Some proteins, such as the displayed hemoglobin molecule, have more than one polypeptide chain that associate to form the functional unit of the protein; this is called . Questions based upon these scenes: What is the primary sequence shown in the first link? Is the secondary structure shown an alpha helix or beta sheet? The ith C=O of the backbone is hydrogen bonded to which N(-H) (use i +/- # to represent the number)? What atom does this program NOT show? What color is used to represent alpha helices? How many alpha helices are present in the single peptide chain shown? How many polypeptide chains make up the quaternary structure?
Ways of representing protein structureProtein structures can be displayed in many different ways. In models, all of the non-hydrogen atoms are shown as spheres with their van der Waals radii. In the model, the atoms are shown as smaller balls, connected by sticks; this is further simplified in the model, which only shows the bonds between atoms. shows only the N-Calpha-C=O repeating unit; the representation shows the secondary structures. Questions based upon these scenes: Which of these representations would be best for showing... --the secondary structures present in a molecule? --Channels, holes, or pockets in a protein? --Residues in the active site of an enzyme? Explain your answers. Secondary StructuresIn this section, you will both learn about secondary structure properties and manipulating structures in Jmol. We will begin with some basic manipulation strategies so that you can analyze secondary structures. Try the following manipulations with the mouse: --Click and move the mouse to the right, the left, up, and down; what happens to the molecule? --Hold the shift button while you try the same manipulations. What does each do? Clicking the right mouse button in the structure box brings up an extensive menu. This exercise will use commands in the style, color,zoom, measurements, and set picking categories. We will begin with the structure of an alpha helix from hemoglobin. From this view, can you determine: --The number of amino acids per turn? --The position of the side chains? Hold the mouse over each end of the alpha helix. A yellow box should appear, with [VAL]17:A:CA:#120. This indicates the amino acid residue, the position in the chain, which chain, what atom it is (CA means the alpha carbon), and the overall number of the atom. If there are two identical chains, one of the chains may be numbered slightly differently (like adding 200 to each residue number) to distinguish the residues. --What is the amino acid range (numbers) of this alpha helix? Rotate the helix so that you are looking down the helix. What does the middle of the helix look like? Right click on the mouse, choose style, then scheme, then CPK spacefill. What does the middle of the helix look like? Which view is more representative of the true structure of the molecule? Let's try changing to another view. Right click on the mouse, choose style, then scheme, then ball and stick. Based upon what you know about peptide composition or by holding the mouse over the atoms determine the color scheme: red = black = blue = Notice that hydrogens are not shown on this model. Xray crystallography is not able to resolve hydrogens, so they are omitted from the images. This also simplifies the data set, as there are many fewer atoms to position. From this view, it is possible to determine the number of amino acids in one turn. has the side chains removed (though the alpha carbons show where the side chain would be).
|
| ||||||||||