Nitric Oxide Synthase
Introduction to NOSIntroduction to NOS
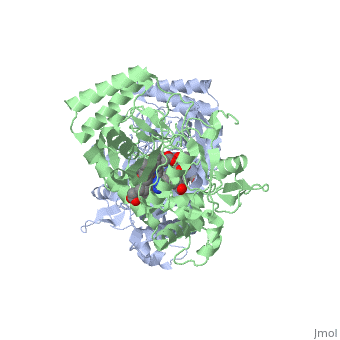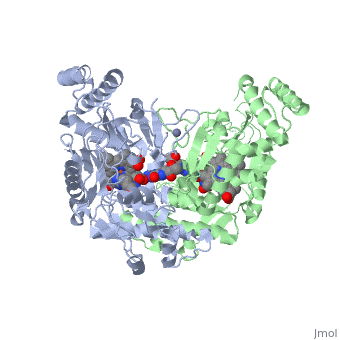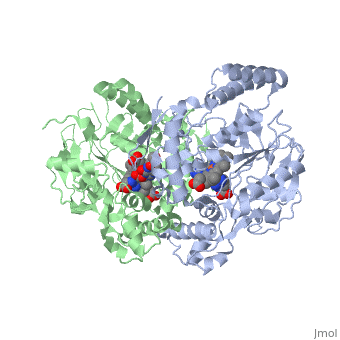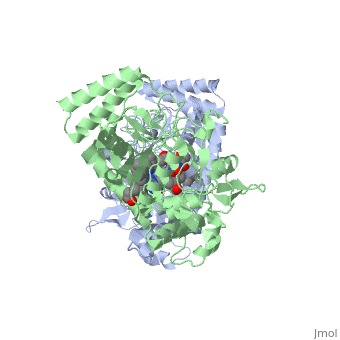
Nitric Oxide Synthase (NOS) is a group of enzymes catalysing L-arginine to yield L-Citrulline and Nitric Oxide[1] (NO). NOS is a homodimeric protein with 125- to 160-kD subunits. An overview of the NOS homodimer is given below. All cofactors are included and the electron transfer pathway which takes place in NOS is indicated.
The NOS homodimer is composed of two subunits, each containing two domains: an oxygenase domain and a reductase domain. The subunits are held together by a Zinc ion, which is bound by 4 cystein amino acid present in the oxygenase domain, two in each domain. Further, many amino acid interactions also hold the sunbunits together. Binding of the two types of domains is caused by CaM. The reductase domain supplies electrons for the NOS reaction which takes place in the oxygenase domain. The reductase domain contains two redox-active prosthetic groups, FAD and FMN. NADPH binds to the domain and passes on an electron to FAD which passes the electron on to FMN. FMN passes the electron on to the Heme in the oxygenase domain of the opposite subunit. The oxygenase domain contains H4B (5,6,7,8-tetrahydrobiopterin)and the already mentioned Heme ion (Fe(III)). These two are also redox active groups. H4B is required by NOS in order to produce NO and not H2O2. Besides Heme and H4B, the oxygenase domain binds the substrate L-arginine which takes part in the NO synthase reaction (see below).
In mammals three isozymes of NOS has been identified: Neuronal NOS (nNOS), inducible NOS (iNOS), and endothelial NOS (eNOS). One differentiates between constitutive NOS (always produced - eNOS and nNOS) and inducable NOS (iNOS). Constitutive NOS are regulated by calcium binding to the CaM region and is thus regulated by calcium. nNOS produces NO in nervous tissue in both the peripheral and the central nervous system. nNOS functions in cell signaling and communication - a vital part of the nervous tissue. eNOS controls the amount of NO signaling in the endothelial cells (eg. blood vessel dilation). iNOS is induced to produce NO only when needed. For example when the immune system is activated. iNos is not regulated by calcium. The NOS enzymes is found in numeral organisms. Most facts used at this page are taken from the human NOS. The active site and different binding regions are highly conserved and therefore sites from other organisms will be used as well.
The reaction of NOSThe reaction of NOS
As previously described NOS is an enzyme split in to different domains; the N-terminal oxygenase domain and the C-terminal reductase domain.
The oxygenase domain is where the production of NO takes place, whereas the reductase domain provides the electrons necessary to drive the reaction in the oxygenase domain. The reaction is:
The amino acid L-Arginine is turned in to L-Citrulline and NO. The reaction is driven by the oxidation of NADPH to NADP+, which in total yields 5 electrons for the reaction. The reaction above therefore takes both the oxygenase and the reductase domain into account.
The Oxygenase domain of NOSThe Oxygenase domain of NOS
The contains the active site of the enzyme. The active site binds the substrate L- (Arginine) which is converted into citruline and NO (explained in details below). The domain has three cofactors bound:
((6R).5,6,7,8-Tetrahydrobiopterin)
(Heme)
and a .
General StructureGeneral Structure
The oxygenase domain can be divided into three subdomains. Firstly, the substrate-binding subdomain, which is crescent in shape, binds the substrate in an interior pocket of the crescent. The Heme group is located between the tips of the crescent shape, thus closing the pocket in which the substrate is located. Secondly the the H4B binding subdomain serves as a cap for the for the cavity created by the substrate binding domain's crescent shape. Thirdly there is a subdomain with two helical bundles making up a hydrophobic core, however this subdomain is not thought to take part of the catalytic activity of the enzyme.
Substrate bindingSubstrate binding
The active site is highly conserved in the different NOS species. Thus it is possible to discuss substrate binding i general terms. The NOS enzyme binds its substrate by hydrogen bindings both to the guanidino[2] end and the amino acid end (EVT FIG!).
binds its substrate (L-arginine) by coordinating CO to the heme at the site occupied by oxygen....[1].
H4BH4B
|
is a cofactor. NOS contains two molecules of , one in each monomer. The active center forms a kind of tunnel, because of the dimeric structure. This gives H4B the opportunity to play a big role in the control of subunit interactions and active-center formation. H4B therefor is more of a structurel cofactor, in that it keeps the dimer stabilized by integration in to the hydrophobic parts of the dimer. Here it helps substrate interactions by lining the active-center channel and hydrogen bonding to the heme propionate amd to alfa7 which is two elements involved in L-Arg binding. Its structural importense is also reconned to play a role in dimer formation, and major conformational changes leading to the formation af the active site channelform.[2]. The H4B is bound by hydrogen-bonds to several of the molekules surrounding it, including the substrate L-Arg. The substrate is H-bonded to the 4-keto group of pterin, and to one of the heme propionate groups, that has two carboxylate oxygens in use for H-bonds. These oxygens are further H-bonded to the 4-keto group of pterin, through water, and directly to N(3) and NH2 on C (2). The big picture of all the H-bonds can be seen on figure (???)-lav figur i chemdraw inspireret af figuren s. 943Raman)))
But H4B is not only a structurel cofactor, it also plays a very important role in NO synthesis, donating an electron to the heme.[3] H4B can deliver an electron to the heme much faster than the reductase domain can, therefor H4B is used by NOS in the Arg hydroxylation, activating O2 by providing the second electron. So H4B is a kinetically prefered electron donor.
(indsæt fig.3 i artiklen) As shown in the reaction (fig.3) the second electron, that H4B donates helps the FeIIO2 intermadiate to be reduced in to oxidants that can react with Arg and N-hydroxy-L-arginine (NOHA) [4] If H4B was not present the FeIIO2 intermediate would decay to superoxide and ferric enzyme, because the reductase domain is slower to deliver an electron, than the proces of decay is to happen. But H4B is faster than both of these processes. [5]
HemeHeme
ZincZinc
The Reductase Domain of NOSThe Reductase Domain of NOS
Template:STRUCTURE 1tll The reductase domain of the NOS homodimer will not be discussed thoroughly at this page. However a short discussion of the electron transfer which occurs will be given along with an introduction to the bound cofactors and the general structure.
The has three cofactors bound, here is only shown one subunit of the domain:
(Nicotinamide adenine dinucleotide phosphate)
The reductase domain is, as mentioned, bound to an oxygenase domain by a calmodulin linker. This linker responds to Ca2+ -ions (constitutive NOS isoforms). The calmodulin linker is consists of 32 residues and contains a binding region for the Ca2+-ions. This binding is found to be crucial it induces a conformational change which is essential for the electron transfer. It is important to emphasize that the electron transfer occurs from the reductase domain of one subunit to the oxygenase domain of the opposite subunit (i.e. a trans transfer). The conformational change induced by Ca2+-ions brings the mentioned reductase and oxygenase domains closer together, therefore the linker acts like a hinge. The electron transfer occurs two times per produced NO molecule, first electrons are passed on for the conversion of L-Arginine to its intermediate, secondly for the conversion of the intermediate to produce Citruline and NO. In general the reductase domain can be divided into three binding domains: the NADPH binding domain, the FAD binding domain, and the FMN binding domain. The NADPH and FAD binding domains are associated whereas the FAD and FMN domains are connected by an α-helical binding domain. An electron is donated by NADPH, which passes the electron on to FAD. FAD shuttles on the electron to FMN. The FMN binding domain is a flexible domain and here the conformational change occurs, the Calmodulin linker rotates the reductase domain and oxygenase domain along a vertical axis, thus bringing the reductase domain closer to the opposite oxygenase domain. The electron can then due to shorter distance be passed on the the Heme group bound by the oxygenase domain [6].
ReferencesReferences
- ↑ Fan B, Wang J, Stuehr DJ, Rousseau DL. NO synthase isozymes have distinct substrate binding sites. Biochemistry. 1997 Oct 21;36(42):12660-5. PMID:9376373 doi:http://dx.doi.org/10.1021/bi9715369
- ↑ Raman CS, Li H, Martasek P, Kral V, Masters BS, Poulos TL. Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center. Cell. 1998 Dec 23;95(7):939-50. PMID:9875848
- ↑ Wei CC, Wang ZQ, Meade AL, McDonald JF, Stuehr DJ. Why do nitric oxide synthases use tetrahydrobiopterin? J Inorg Biochem. 2002 Sep 20;91(4):618-24. PMID:12237227
- ↑ Wei CC, Wang ZQ, Meade AL, McDonald JF, Stuehr DJ. Why do nitric oxide synthases use tetrahydrobiopterin? J Inorg Biochem. 2002 Sep 20;91(4):618-24. PMID:12237227
- ↑ Wei CC, Wang ZQ, Meade AL, McDonald JF, Stuehr DJ. Why do nitric oxide synthases use tetrahydrobiopterin? J Inorg Biochem. 2002 Sep 20;91(4):618-24. PMID:12237227
- ↑ Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J Biol Chem. 2004 Sep 3;279(36):37918-27. Epub 2004 Jun 17. PMID:15208315 doi:http://dx.doi.org/10.1074/jbc.M406204200