1ykf
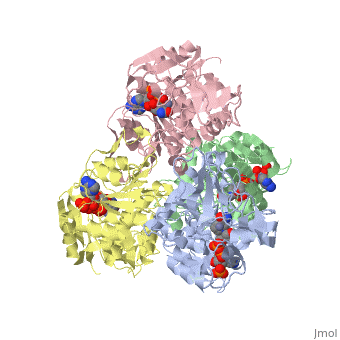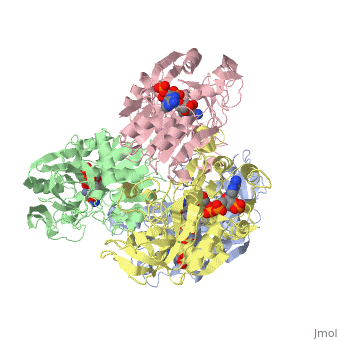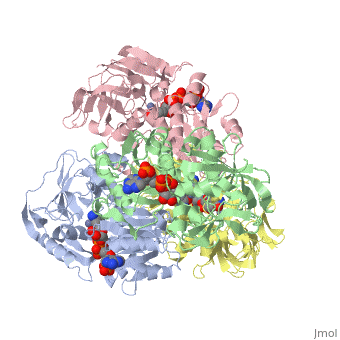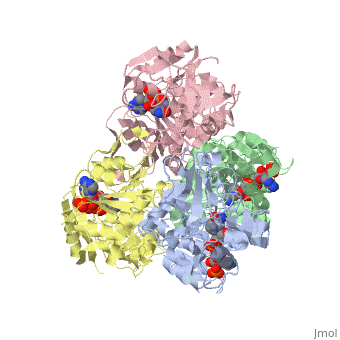
NADP-DEPENDENT ALCOHOL DEHYDROGENASE FROM THERMOANAEROBIUM BROCKIINADP-DEPENDENT ALCOHOL DEHYDROGENASE FROM THERMOANAEROBIUM BROCKII
Structural highlights
Function[ADH_THEBR] Alcohol dehydrogenase with a preference for medium chain secondary alcohols, such as 2-butanol and isopropanol. Has very low activity with primary alcohols, such as ethanol. Under physiological conditions, the enzyme reduces aldehydes and 2-ketones to produce secondary alcohols. Is also active with acetaldehyde and propionaldehyde. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedWe have determined the X-ray structures of the NADP(H)-dependent alcohol dehydrogenase of Clostridiim beijerinckii (CBADH) in the apo and holo-enzyme forms at 2.15 A and 2.05 A resolution, respectively, and of the holo-alcohol dehydrogenase of Thermoanaerobacter brockii (TBADH) at 2.5 A. These are the first structures of prokaryotic alcohol dehydrogenase to be determined as well as that of the first NADP(H)-dependent alcohol dehydrogenase. CBADH and TBADH 75% have sequence identity and very similar three-dimensional structures. Both are tetramers of 222 symmetry. The monomers are composed of two domains: a cofactor-binding domain and a catalytic domain. These are separated by a deep cleft at the bottom of which a single zinc atom is bound in the catalytic site. The tetramers are composed of two dimers, each structurally homologous to the dimer of alcohol dehydrogenases of vertebrates. The dimers form tetramers by means of contacts between surfaces opposite the interdomain cleft thus leaving it accessible from the surface of the tetramer. The tetramer encloses a large internal cavity with a positive surface potential. A molecule of NADP(H) binds in the interdomain cleft to the cofactor-binding domain of each monomer. The specificity of the two bacterial alcohol dehydrogenases toward NADP(H) is determined by residues Gly198, Ser199, Arg200 and Tyr218, with the latter three making hydrogen bonds with the 2'-phosphate oxygen atoms of the cofactor. Upon NADP(H) binding to CBADH, Tyr218 undergoes a rotation of approximately 120 degrees about chi1 which facilitates stacking interactions with the adenine moiety and hydrogen bonding with one of the phosphate oxygen atoms. In apo-CBADH the catalytic zinc is tetracoordinated by side-chains of residues Cys37, His59, Asp150 and Glu60; in holo-CBADH, Glu60 is retracted from zinc in three of the four monomers whereas in holo-TBADH, Glu60 does not participate in Zn coordination. In both holo-enzymes, but not in the apo-enzyme, residues Ser39 and Ser113 are in the second coordination sphere of the catalytic zinc. The carboxyl group of Asp150 is oriented with respect to the active carbon of NADP(H) so as to form hydrogen bonds with both pro-S and pro-R hydrogen atoms. NADP-dependent bacterial alcohol dehydrogenases: crystal structure, cofactor-binding and cofactor specificity of the ADHs of Clostridium beijerinckii and Thermoanaerobacter brockii.,Korkhin Y, Kalb(Gilboa) AJ, Peretz M, Bogin O, Burstein Y, Frolow F J Mol Biol. 1998 May 22;278(5):967-81. PMID:9836873[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||