1a0a
PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEXPHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX
Structural highlights
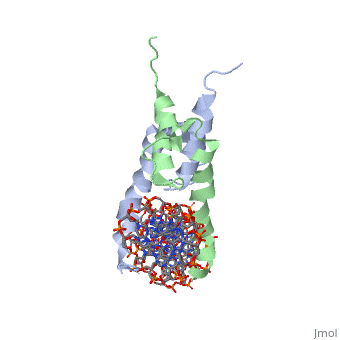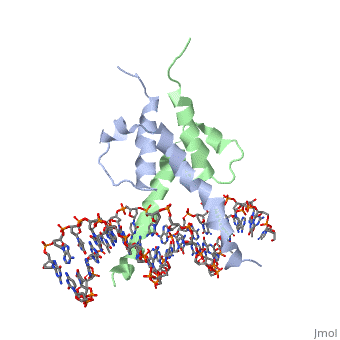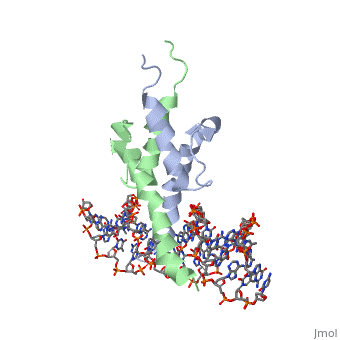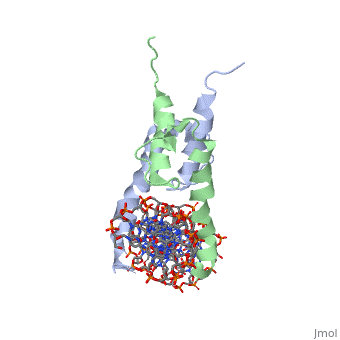
Function[PHO4_YEAST] Transcriptional activator that regulates the expression of repressible phosphatase under phosphate starvation conditions. Binds to the upstream activating sequence (UAS) of several phosphatase encoding PHO genes. Inhibited by the cyclin-CDK PHO80-PHO85 under high-phosphate conditions. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe crystal structure of a DNA-binding domain of PHO4 complexed with DNA at 2.8 A resolution revealed that the domain folds into a basic-helix-loop-helix (bHLH) motif with a long but compact loop that contains a short alpha-helical segment. This helical structure positions a tryptophan residue into an aromatic cluster so as to make the loop compact. PHO4 binds to DNA as a homodimer with direct reading of both the core E-box sequence CACGTG and its 3'-flanking bases. The 3'-flanking bases GG are recognized by Arg2 and His5. The residues involved in the E-box recognition are His5, Glu9 and Arg13, as already reported for bHLH/Zip proteins MAX and USF, and are different from those recognized by bHLH proteins MyoD and E47, although PHO4 is a bHLH protein. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.,Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T EMBO J. 1997 Aug 1;16(15):4689-97. PMID:9303313[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||