1shy
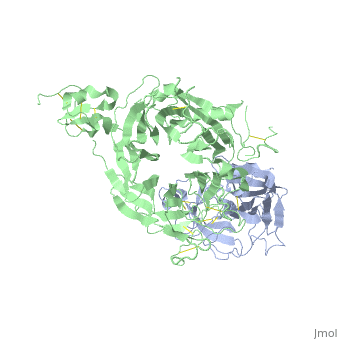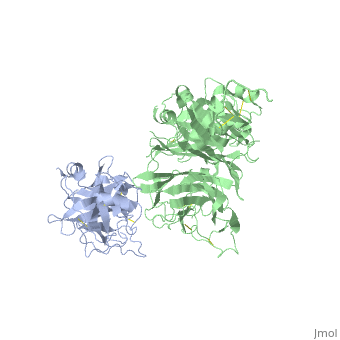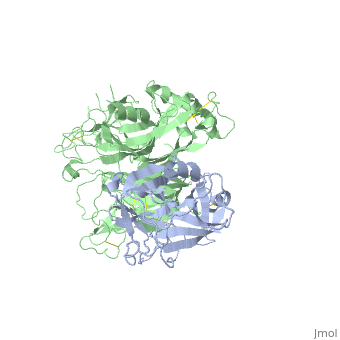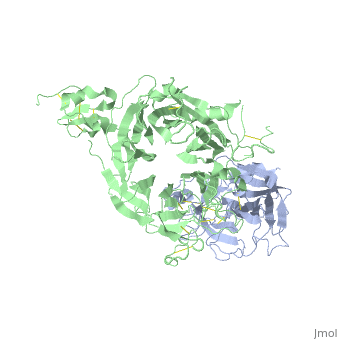
The Crystal Structure of HGF beta-chain in Complex with the Sema Domain of the Met Receptor.The Crystal Structure of HGF beta-chain in Complex with the Sema Domain of the Met Receptor.
Structural highlights
Disease[HGF_HUMAN] Defects in HGF are the cause of deafness autosomal recessive type 39 (DFNB39) [MIM:608265]. A form of profound prelingual sensorineural hearing loss. Sensorineural deafness results from damage to the neural receptors of the inner ear, the nerve pathways to the brain, or the area of the brain that receives sound information.[1] [MET_HUMAN] Note=Activation of MET after rearrangement with the TPR gene produces an oncogenic protein. Note=Defects in MET may be associated with gastric cancer. Defects in MET are a cause of hepatocellular carcinoma (HCC) [MIM:114550].[2] Defects in MET are a cause of renal cell carcinoma papillary (RCCP) [MIM:605074]. It is a subtype of renal cell carcinoma tending to show a tubulo-papillary architecture formed by numerous, irregular, finger-like projections of connective tissue. Renal cell carcinoma is a heterogeneous group of sporadic or hereditary carcinoma derived from cells of the proximal renal tubular epithelium. It is subclassified into common renal cell carcinoma (clear cell, non-papillary carcinoma), papillary renal cell carcinoma, chromophobe renal cell carcinoma, collecting duct carcinoma with medullary carcinoma of the kidney, and unclassified renal cell carcinoma.[3] [4] [5] [6] [7] Note=A common allele in the promoter region of the MET shows genetic association with susceptibility to autism in some families. Functional assays indicate a decrease in MET promoter activity and altered binding of specific transcription factor complexes. Note=MET activating mutations may be involved in the development of a highly malignant, metastatic syndrome known as cancer of unknown primary origin (CUP) or primary occult malignancy. Systemic neoplastic spread is generally a late event in cancer progression. However, in some instances, distant dissemination arises at a very early stage, so that metastases reach clinical relevance before primary lesions. Sometimes, the primary lesions cannot be identified in spite of the progresses in the diagnosis of malignancies.[8] Function[HGF_HUMAN] Potent mitogen for mature parenchymal hepatocyte cells, seems to be a hepatotrophic factor, and acts as a growth factor for a broad spectrum of tissues and cell types. Activating ligand for the receptor tyrosine kinase MET by binding to it and promoting its dimerization.[9] [10] [MET_HUMAN] Receptor tyrosine kinase that transduces signals from the extracellular matrix into the cytoplasm by binding to hepatocyte growth factor/HGF ligand. Regulates many physiological processes including proliferation, scattering, morphogenesis and survival. Ligand binding at the cell surface induces autophosphorylation of MET on its intracellular domain that provides docking sites for downstream signaling molecules. Following activation by ligand, interacts with the PI3-kinase subunit PIK3R1, PLCG1, SRC, GRB2, STAT3 or the adapter GAB1. Recruitment of these downstream effectors by MET leads to the activation of several signaling cascades including the RAS-ERK, PI3 kinase-AKT, or PLCgamma-PKC. The RAS-ERK activation is associated with the morphogenetic effects while PI3K/AKT coordinates prosurvival effects. During embryonic development, MET signaling plays a role in gastrulation, development and migration of muscles and neuronal precursors, angiogenesis and kidney formation. In adults, participates in wound healing as well as organ regeneration and tissue remodeling. Promotes also differentiation and proliferation of hematopoietic cells.[11] [12] [13] Acts as a receptor for Listeria internalin inlB, mediating entry of the pathogen into cells.[14] [15] [16] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe Met tyrosine kinase receptor and its ligand, hepatocyte growth factor (HGF), play important roles in normal development and in tumor growth and metastasis. HGF-dependent signaling requires proteolysis from an inactive single-chain precursor into an active alpha/beta-heterodimer. We show that the serine protease-like HGF beta-chain alone binds Met, and report its crystal structure in complex with the Sema and PSI domain of the Met receptor. The Met Sema domain folds into a seven-bladed beta-propeller, where the bottom face of blades 2 and 3 binds to the HGF beta-chain 'active site region'. Mutation of HGF residues in the area that constitutes the active site region in related serine proteases significantly impairs HGF beta binding to Met. Key binding loops in this interface undergo conformational rearrangements upon maturation and explain the necessity of proteolytic cleavage for proper HGF signaling. A crystallographic dimer interface between two HGF beta-chains brings two HGF beta:Met complexes together, suggesting a possible mechanism of Met receptor dimerization and activation by HGF. Crystal structure of the HGF beta-chain in complex with the Sema domain of the Met receptor.,Stamos J, Lazarus RA, Yao X, Kirchhofer D, Wiesmann C EMBO J. 2004 Jun 16;23(12):2325-35. Epub 2004 May 27. PMID:15167892[17] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||