1d5f
STRUCTURE OF AN E6AP-UBCH7 COMPLEX: INSIGHTS INTO THE UBIQUITINATION PATHWAY
| |||||||
| , resolution 2.8Å | |||||||
|---|---|---|---|---|---|---|---|
| Coordinates: | save as pdb, mmCIF, xml | ||||||
OverviewOverview
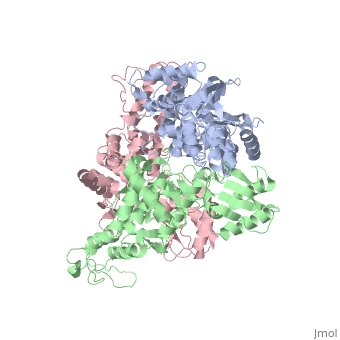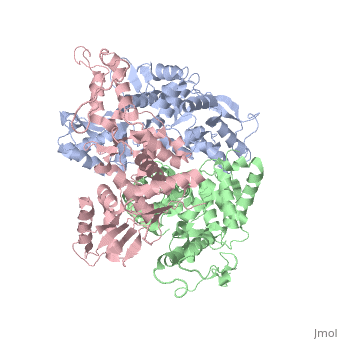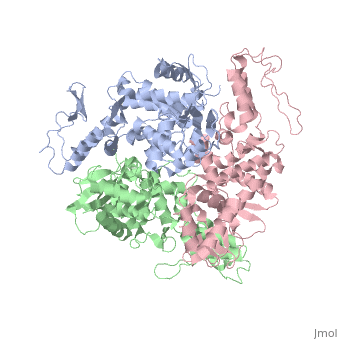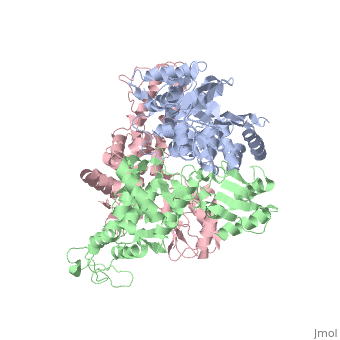
The E6AP ubiquitin-protein ligase (E3) mediates the human papillomavirus-induced degradation of the p53 tumor suppressor in cervical cancer and is mutated in Angelman syndrome, a neurological disorder. The crystal structure of the catalytic hect domain of E6AP reveals a bilobal structure with a broad catalytic cleft at the junction of the two lobes. The cleft consists of conserved residues whose mutation interferes with ubiquitin-thioester bond formation and is the site of Angelman syndrome mutations. The crystal structure of the E6AP hect domain bound to the UbcH7 ubiquitin-conjugating enzyme (E2) reveals the determinants of E2-E3 specificity and provides insights into the transfer of ubiquitin from the E2 to the E3.
DiseaseDisease
Known disease associated with this structure: Angelman syndrome OMIM:[601623]
About this StructureAbout this Structure
1D5F is a Single protein structure of sequence from Homo sapiens. Full crystallographic information is available from OCA.
ReferenceReference
Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade., Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP, Science. 1999 Nov 12;286(5443):1321-6. PMID:10558980
Page seeded by OCA on Thu Mar 20 10:33:08 2008