Motifs In ProteinsMotifs In Proteins
The term "motif" when used in structural biology tends to refer to one of two cases:
- A particular amino-acid sequence that characterises a biochemical function
- A set of secondary structure elements that defines a functional or structural role
There are a great number of protein sequence motifs identified, many of which have well defined structural or functional roles. One such example of this is the so-called zinc finger motif which is readily identified from the following consensus sequence pattern (where "X" represents any amino acid):
Cys - X(2-4) - Cys - X(3) - Phe - X(5) - Leu - X(2) - His - X(3) - His
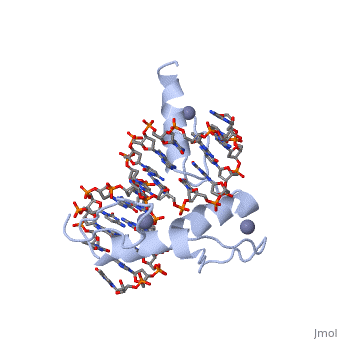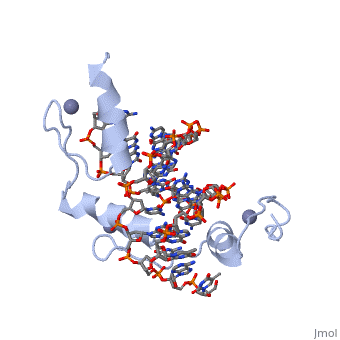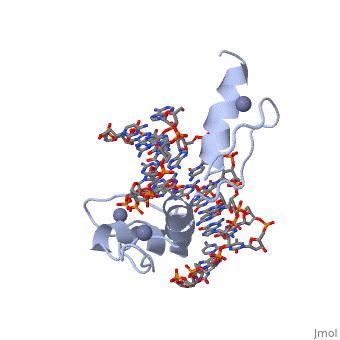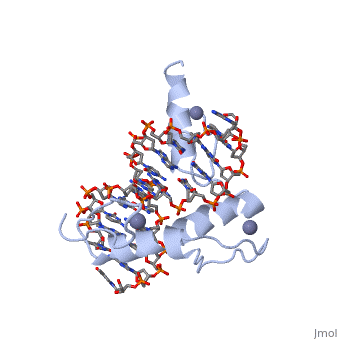
The example structure shown to is that of Zif268 protein-DNA complex from Mus musculus (PDB entry 1AAY). In this example (a C2H2 class zinc finger) the conserved and residues form ligands to a whose coordination is essential to stabilise the tertiary fold of the protein. The fold is important because it helps orientate the to bind to the .
However, there are also a number of repeated patterns and functional motifs revealed when the protein structure is examined. This page aims to introduce some of the main types of motif illustrating them on protein structures from the PDB.
Secondary structure elementsSecondary structure elements