Sandbox SouthUniversity1
==Selectivity of Drug Metabolism by Cytochrome P450 Enzymes==
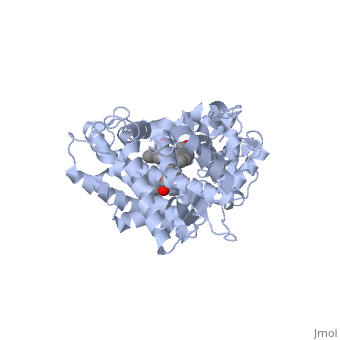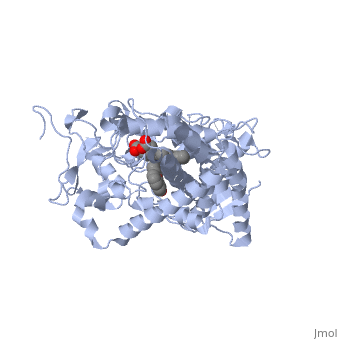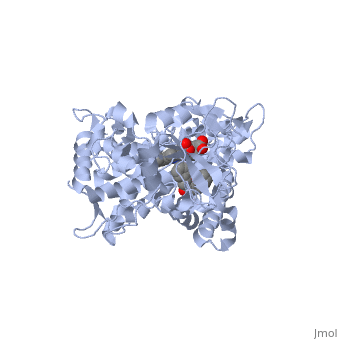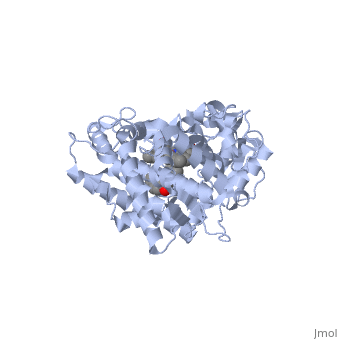
This tutorial covers the basic structure and function of the Cytochrome P450 (CYP) enzymes, the most important group of drug metabolizing enzymes. The example used here for illustration purposes is the second protein discovered in the CYP 1 family, in subfamily A (generally referred to as CYP1A2). This protein is shown in an interactive window to the right. Turn off (toggle) spinning of the protein by clicking on the button below the structure. The quality of the molecule rendering can also be increased by clicking the "toggle quality" button, although this may decrease the smoothness when the molecule is rotating. Now rotate the molecule by clicking and dragging in the window with your cursor. Resize the molecule by holding down the shift key and dragging up and down. Rotate and resize the molecule until you can clearly see that there are 2 molecules shown in a "space-filling" representation in the middle of the protein (they are more or less perpendicular to each other, and almost touching). These are a heme molecule, which is absolutely vital for the enzyme's function, and a second molecule (alpha-naphthoflavone) which represents a compound that is about to be metabolized. First, lets look at the heme. Click the following link to hide the rest of the protein and Carbon atoms are shown in grey, Nitrogen atoms are blue, Oxygen is red, and the central iron atom is orange. The iron atom is a vital center for oxidation of substrates (drugs or other xenobiotics). It is shown with the protein strands displayed partially transparent to highlight the heme ring system and iron atom. Now look at the substrate molecule being oxidized and view its orientation relative to the heme group by clicking Resize and rotate the molecules until you can see how the two molecules are oriented in relationship to each other. The flavone is metabolized (oxidized) by introduction of a hydroxy group onto the phenyl ring that is attached to the tricyclic ring system.
Purely based on the distance between the pheny ring of the flavone substrate and the heme iron, oxidation might be expected to take place at the 4 (para) position. However, other factors also help determine the regioselectivity of the metabolism (selectivity for oxidation at the different possible positions). One of these factors is the relative reactivity of the various positions on the substrate.
|
| ||||||||||
Different members of the CYP450 enzymes preferentially metabolize different xenobiotics. What features of a drug do you think might cause it to be metabolized by one CYP versus another?
Draft: <illustration of residues in active site>
Draft animation
Draft animation
draft: <insert quiz here on electrostatic and steric factors>
draft: <insert animations here contrasting the shape of the active site of CYP3A4 and e.g. CYP2E1>
draft: <insert examples of substrates for 2 different CYPs, overlay them on top of each other illustrate the molecular commonalities>
draft <illustrate the size of the binding pockets and their electronic character>
draft test of scene names:
<Draft Quiz>
References The structure of CYP1A2 shown above is found in the Research Collaboration of Structural Bioinformatics Protein Database as entry 2HI4.