Conservation, Evolutionary: Difference between revisions
Eric Martz (talk | contribs) →Limiting ConSurf Analysis to Proteins of a Single Function: revised for changes at ConSurf |
Eric Martz (talk | contribs) linked 'taxa' to wikipedia |
||
| Line 1: | Line 1: | ||
Mutations occur spontaneously in each generation, randomly changing the amino acid sequences of proteins. Individuals with mutations that impair critical functions of proteins may have resulting problems that make them less able to reproduce. Harmful mutations are lost from the gene pool because the individuals carrying them reproduce less effectively. Over time, only harmless (or very rare beneficial) mutations are maintained in the gene pool. This is [[Evolution|evolution]]. | Mutations occur spontaneously in each generation, randomly changing the amino acid sequences of proteins. Individuals with mutations that impair critical functions of proteins may have resulting problems that make them less able to reproduce. Harmful mutations are lost from the gene pool because the individuals carrying them reproduce less effectively. Over time, only harmless (or very rare beneficial) mutations are maintained in the gene pool. This is [[Evolution|evolution]]. | ||
When the sequences of a given protein are compared between taxa, using multiple sequence alignment (MSA), differences between sequences most often represent mutations that were allowed (by evolution) to persist because they were harmless. Where the sequences are identical, we say that sequence was '''conserved'''. Such '''evolutionary conservation''' occurs because mutations of these amino acids were harmful to protein function, and were lost over time. Amino acids that are conserved are those most critical to the function of the protein. Thus, looking for evolutionarily conserved patches of amino acids in a 3D protein structure is a good way to '''locate functional sites'''. | When the sequences of a given protein are compared between [http://en.wikipedia.org/wiki/Taxa taxa], using multiple sequence alignment (MSA), differences between sequences most often represent mutations that were allowed (by evolution) to persist because they were harmless. Where the sequences are identical, we say that sequence was '''conserved'''. Such '''evolutionary conservation''' occurs because mutations of these amino acids were harmful to protein function, and were lost over time. Amino acids that are conserved are those most critical to the function of the protein. Thus, looking for evolutionarily conserved patches of amino acids in a 3D protein structure is a good way to '''locate functional sites'''. | ||
{| class="wikitable" width="600" align="right" | {| class="wikitable" width="600" align="right" | ||
Revision as of 09:04, 4 June 2009
Mutations occur spontaneously in each generation, randomly changing the amino acid sequences of proteins. Individuals with mutations that impair critical functions of proteins may have resulting problems that make them less able to reproduce. Harmful mutations are lost from the gene pool because the individuals carrying them reproduce less effectively. Over time, only harmless (or very rare beneficial) mutations are maintained in the gene pool. This is evolution.
When the sequences of a given protein are compared between taxa, using multiple sequence alignment (MSA), differences between sequences most often represent mutations that were allowed (by evolution) to persist because they were harmless. Where the sequences are identical, we say that sequence was conserved. Such evolutionary conservation occurs because mutations of these amino acids were harmful to protein function, and were lost over time. Amino acids that are conserved are those most critical to the function of the protein. Thus, looking for evolutionarily conserved patches of amino acids in a 3D protein structure is a good way to locate functional sites.

|
The nine conservation grade colors utilized by ConSurf-DB and ConSurf, plus yellow for amino acids with insufficient data, and gray for chains that ConSurf could not process. See Help:Color Keys. |
| |
Locating Conserved PatchesLocating Conserved Patches
Patches of highly conserved amino acid residues on the surface of a protein molecular structure are good candidates for functional sites. Every article in Proteopedia that is titled with a PDB code has an Evolutionary Conservation section below the molecular scene. Clicking show in the blue Evolutionary Conservation bar automatically colors all chains in the molecule by evolutionary conservation as calculated by ConSurf-DB.
Briefly, ConSurf-DB gathers sequences similar to that of the protein in question, then constructs a multiple sequence alignment, and analyses it for sequence positions that are conserved (have lower than average differences between sequences) and that are variable (have higher than average differences between sequences). Each amino acid is assigned a conservation score and corresponding color in Proteopedia's interactive 3D molecular scene.
ConSurf-DB's analysis is done with sophisticated, published, peer-reviewed, state of the art methods. A more detailed overview of the mechanism employed by ConSurf-DB is summarized below. Proteopedia's built-in display of ConSurf-DB results is a good place to start looking for conserved patches.
However, as explained below, ConSurf-DB usually does not show all the conserved patches present in proteins with the same function. Therefore, you may wish to extend your analysis of conservation by limiting the analysis to proteins of one function, using the ConSurf Server, as explained below. The results of such an analysis can be displayed in a molecular scene in Proteopedia. See below for Examples and Instructions.
Topic pages in Proteopedia (manually-authored pages that typically discuss more than one PDB code) may include molecular scenes colored by evolutionary conservation. See below for Examples and Instructions.
Locating Variable PatchesLocating Variable Patches
In some cases, patches of highly variable (rapidly mutating) residues are also functional sites. These can also be identified with Proteopedia's Evolutionary Conservation scenes. For example, mutations in influenza hemagglutinin help the virus to evade host defenses (see 1hgf). Another example is the high allelic variability of the peptide-binding groove of Major Histocompatibility Complex Class I. That variability helps the grooves of the alleles within any individual to bind a wide range of peptides, hence enabling the T lymphocyte system to defend against a wide range of pathogens, including influenza virus. See the ConSurf-colored example below.
Conservation for Domain FoldingConservation for Domain Folding
Certain residues on the surfaces of protein molecules tend to be conserved in order to maintain proper folding, rather than because they are part of a site functioning to interact with substrate, ligand, or a protein partner. Secondary structure elements need to break at the protein molecular surface in order to turn back into the folded protein domain. Therefore, it is common to see isolated highly conserved residues that enable turns, or break helices, notably glycines or prolines, on protein structure surfaces.
Remember that you can touch any residue with the mouse in the Evolutionary Conservation scene in Proteopedia (in Jmol), and its identity will be displayed after a few seconds. This works best with spinning turned off.
Every structure in Proteopedia has a link to be displayed in FirstGlance in Jmol. There, you can use the Find dialog to enter the name of an amino acid, e.g. glycine or proline, and the positions of all of the specified amino acids will be highlighted. You can then visualize their distribution in the 3D structure.
CaveatsCaveats
ConSurf-DB Often Obscures Some Functional SitesConSurf-DB Often Obscures Some Functional Sites
Proteopedia's Evolutionary Conservation scenes use pre-calculated results from ConSurf-DB. ConSurf-DB is designed to include a wide range of sequences in its multiple-sequence alignments (MSA) and analyses. Often, the MSA will a include substantial number of sequences for proteins with different functions than the query protein. (See below for how to find out the functions of the proteins used in ConSurf-DB's MSA.) Consequently, amino acids that are colored as highly conserved by ConSurf-DB are truly highly conserved across a wide range of sequence-similar proteins. However, amino acids that are highly conserved in proteins with the same function as the query protein may not appear conserved in ConSurf-DB results. A good way to find these obscured functional sites is to do a conservation analysis that is limited to proteins of a single function. See below for instructions.
Use Caution When Comparing Conservation of Sequence-Different ChainsUse Caution When Comparing Conservation of Sequence-Different Chains
This caveat applies only to molecules that contain chains with different sequences. The conservation colors shown in Proteopedia's Evolutionary Conservation scenes do not indicate the same levels of conservation for chains of different sequences. This is because ConSurf-DB calculates conservation levels independently for each sequence-different chain, and the levels are relative to the multiple sequence alignment constructed for each sequence-independent chain.
For example, consider 1bqh, which contains 10 chains, representing two copies of a 5-chain molecule. Each molecule contains four sequence-different chains. A visit to ConSurf-DB reveals, as expected, that a different number of sequences was utilized for the multiple sequence alignment (MSA) and conservation calculations for each of these sequence-different chains, and that each MSA had a different average pairwise difference (APD), a measure of diversity within the MSA. Therefore, residues with, for example, conservation level 9 (maximal conservation) in each of the three ConSurf-DB-colored sequence-different chains have the highest levels of conservation within their own chain, but do not have exactly the same absolute levels of conservation.
| 1bqh | |||
| Chain | Length | Number of sequences in MSA | APD |
|---|---|---|---|
| A | 274 | 144 | 1.72 |
| B | 99 | 75 | 1.49 |
| C | 8 | Length below minimum for ConSurf | |
| G | 129 | 201 | 1.35 |
In Proteopedia's Evolutionary Conservation scenes, all the chains in the molecule are colored in the same scene. This gives a potentially useful overview, but can be misleading unless one realizes that a given conservation color, in two sequence-different chains, does not mean exactly the same level of conservation. In contrast to Proteopedia's Evolutionary Conservation scenes, ConSurf-DB and ConSurf Server apply conservation level colors to only one chain sequence at a time, thereby avoiding this possible confusion.
Conservation Results Will Change With TimeConservation Results Will Change With Time
Slight variations in the conservation pattern will occur over time, as the number of sequences in the sequence databases used by ConSurf-DB increase. Each update of ConSurf-DB uses somewhat larger sequence databases, and consequently, the MSA's for each chain will be slightly different.
For the same reasons, results from the ConSurf Server will also change slightly with time, even when the job parameters are the same. Only if you upload the same MSA will the results be identical for a given chain when the jobs are run months or years apart.
Examining Functions of Proteins in ConSurf-DB's MSAExamining Functions of Proteins in ConSurf-DB's MSA
As explained above, ConSurf-DB typically includes proteins with more than one function in its conservation analysis. Before deciding whether to do a ConSurf Server job that limits the analysis to proteins of a single function, you may want to see what proteins ConSurf-DB included in its analysis. Here is how to see the names (which hopefully reveal the functions) of the proteins included in ConSurf-DB's analysis of a protein chain. (The following steps are needed in May, 2009. A request to make this easier has been sent to the ConSurf-DB development team.)
- Go to consurfdb.tau.ac.il (the DB, distinct from the ConSurf Server).
- Enter the PDB code (PDB ID) for the protein of interest.
- Click the button for complete results for the chain of interest.
- Under Alignment, note the number of sequences used.
- Under Output Files click on PSI-BLAST output. Download the file seq.blast (OS X) or seq.blast.zip (Windows).
- Windows XP or Vista:
- Double click on seq.blast.zip to unzip it. Right click on seq.blast and Copy. Right click on your Desktop (or elsewhere of your choosing) and Paste. Now you have the unzipped file seq.blast.
- Open seq.blast in a program that can number lines. (Notepad and Wordpad cannot number lines.) Start MS Word or the free Open Office Writer program (available from openoffice.org). Use the File menu to Open seq.blast.
- Delete everything above the first sequence, so the first sequence will be line number 1. The first sequence follows the header Sequences producing significant alignments:.
- Number the sequences by numbering the lines.
- MS Word: search for "add line numbers" to get instructions.
- Open Office Writer: Save the file as seq_blast.txt. (This enables line numbering.) Open the Tools menu, and select Line Numbering....
- Mac OS X:
- In the Finder, right-click (ctrl-click) on the file seq.blast, then Open With an application that can number lines of text. An excellent free one is Textwrangler from BareBones.Com.
- Delete everything above the first sequence, so the first sequence will be line number 1. The first sequence follows the header Sequences producing significant alignments:.
- Number the sequences by numbering the lines.
- MS Word: Set the Open dialog to enable All Files. Search for "add line numbers" to get instructions. You may need to select all and change the font (e.g. to Arial) to get the description of each sequence to fit on one line.
- TextWrangler (or BBEdit): Open the View menu, and under Text Display click Show Line Numbers.
- iWork Pages appears to lack a line numbering capability.
- Now you have the sequences numbered. Find the number equal to the number of sequences used reported under Alignment by ConSurf-DB.
If the functions of the proteins for this sequence number (and lower numbers) differ from that of the protein of interest, then ConSurf-DB included proteins of multiple functions in its analysis. This tends to obscure patches of conservation that exist among proteins with the same function as the query protein of interest.
Limiting ConSurf Analysis to Proteins of a Single FunctionLimiting ConSurf Analysis to Proteins of a Single Function
As explained above, the ConSurf-DB Evolutionary Conservation scene available in Proteopedia often includes proteins with multiple functions. However, the best way to find all functional sites by conservation analysis is to limit the analysis to proteins with a single function. A procedure for doing this follows. In June, 2009, the ConSurf development team is working on a new version that, once released, will enable selection of arbitrary sequences from the PSI-BLAST list.
- Go to consurf.tau.ac.il, the ConSurf Server (distinct from ConSurf-DB).
- Specify your PDB ID, Chain Identifier, and email address.
- Under Advanced Options, set Max. Number of Homologues to all.
- Submit the job.
- When the job is completed, under Running Messages, note the number of unique sequences used in the calculation.
- Under Final Results, Sequences, click on Unique Sequences Used.
- Looking down the list of sequences from the top, find where the function of the protein first differs from that of the query protein of interest. Note the number of the last sequence with the same function as the query protein. We'll call this the max with same function number.
- Re-run your ConSurf job making only one change. Set the Max. Number of Homologues to the "max with same function" that you determined in the previous step.
The results of the final step above may enable you to identify more functional sites than did the ConSurf-DB result built into Proteopedia.
See below for instructions on how to make a green-link scene in Proteopedia that shows your single-function ConSurf result.
If your results have more than a few amino acids with insufficient data (yellow color), you need more sequences. Repeat the procedure above with one change in the ConSurf job submission form: under Advanced Options, use the much larger Uniprot database instead of the default Swiss-Prot database.
The ConSurf-DB MechanismThe ConSurf-DB Mechanism
Because results from the ConSurf DataBase server, ConSurf-DB[1] are displayed within Proteopedia as Evolutionary Conservation, an overview of its methods is provided here. ConSurf-DB pre-calculates conservation levels for each amino acid in every protein chain in the Protein Data Bank. It went into service in 2008. It uses state-of-the-art methods, all published in peer-reviewed journals[1]. Each protein chain is processed as follows.
ConSurf-DB ProcessConSurf-DB Process
- A list of unique protein chains is extracted from the Protein Data Bank. Chains shorter than 30 amino acids are not processed because they do not contain enough information for reliable phylogenetic tree construction. Non-standard residues are converted to the closest standard amino acids. Chains with more than 15% non-standard residues are not processed. Chains that could not be processed are colored gray in Proteopedia -- see the color key at the top of this page.
- The amino acid sequence of each protein chain is submitted to PSI-BLAST[2] for collection of related sequences from UniprotKB/Swiss-Prot[3]. Three iterations are performed using an expectation value[4] cutoff of 10-3.
- The sequences gathered with PSI-BLAST are then filtered (see below) using a scheme that attempts a balance between limiting the sequences to close homologues, and including distant sequences that do not share structure or function.
- The filtered sequence set is multiply aligned with MUSCLE (a multiple sequence alignment algorithm that out-performs CLUSTALW).
- A phylogenetic tree is constructed from the multiple sequence alignment (MSA) using the Rate4Site program developed by the ConSurf team.
- Rate4Site then calculates an evolutionary rate for each position in the MSA using a Bayesian approach shown by the ConSurf team to be superior[5]. "The amino acid evolution is traced using the JTT[6] substitution model. High evolutionary rate represents a variable position while low rate represents an evolutionarily conserved position."[1]
- "The conservation scores are normalized so that the average over all residues is zero, and the standard deviation is one."[1] Thus, conservation scores are relative, not absolute and comparing them between different protein families might be misleading (see Caveat above).
- The normalized conservation scores are then divided into nine levels from 1 (highly variable) to 9 (highly conserved).
- Colors mapped to the nine conservation levels, from turquoise (1) to burgandy (9) are applied to the 3D protein structure visualized in FirstGlance in Jmol. A coloring script for RasMol is also provided.

- A confidence interval for the conservation level is calculated for each amino acid position in the MSA. When this indicates low reliability, the position is colored yellow, signifying that the data were insufficient to assign a meaningful conservation level.
- An Average Pairwise Distance (APD) is calculated to describe the diversity of sequences in the MSA (see below).
The results of each stage of the above process may be viewed for each chain at ConSurf-DB. In the initial run (February 2008), roughly 100 computer CPU's were utilized concurrently via a distributed computing system. Processing of the 30,918 unique protein chains in the PDB took about five days, or an average of roughly 30 minutes per chain.
FilteringFiltering
Filtering of the sequences gathered for each protein chain is crucial to making the ConSurfDB results maximally informative. Filtering consists of the following steps.
- Sequences with more than 95% sequence identity to the query sequence are discarded.
- Sequences shorter than 60% of the query sequence are discarded.
- Locally aligned sequence fragments that overlap by over 10% are discarded.
- Redundant sequences (>95% identical) are removed using CD-HIT[7].
- A maximum of 300 sequences meeting the above criteria is used (the 300 with the lowest expectation values[4], that is, most closely related to the query sequence).
- If the above process yields fewer than 50 sequences, the entire process is repeated using the Clean_UniProt database, which is about ten times larger than UniProtKB/Swiss-Prot. Clean_UniProt is a version of the UniProt database that attempts to exclude mutant or dubious sequences.
- If the above process yields fewer than 5 sequence homologs, no calculation is performed due to insufficient data. In February, 2008, this occurred for 1,348 chains out of 30,918 (4%).
Average Pairwise DistanceAverage Pairwise Distance
An Average Pairwise Distance (APD) is calculated to describe the diversity of sequences in the MSA generated during the processing of each chain. A value of 0.01 means that on average, there is one amino acid replacement for every 100 positions. Optimally informative results are obtained when the APD is between roughly 0.5 and 1.5.
The ConSurf ServerThe ConSurf Server
The ConSurf Server, first available in 2001[8][9][10] with many subsequent enhancements, can calculate and display the conservation pattern for 3D structures completely automatically. Generally, one should use the ConSurf Server only when the pre-calculated result at the ConSurf-DB needs improvement (for example, see above), or if you have your own multiple sequence alignment (MSA) that you wish to use. ConSurf-DB will nearly always give more informative results than the default settings of the ConSurf Server, due to the powerful sequence filtering that is built into ConSurf-DB. For an example, see the cytochrome c comparision at ConSurf-DB.
Like ConSurf-DB, the ConSurf Server uses the same state-of-the-art methods, all of which are published in peer-reviewed journal articles. Unlike ConSurf-DB's pre-calculated results the ConSurf Server permits considerable customization. For example, the user may specify the number of sequences to use, choose the database from which sequences are obtained (Swiss-Prot or UniProt), set the Expectation cutoff[4], set the number of PSI-BLAST iterations, or submit their own multiple sequence alignment, or phylogenetic tree. Also you can upload your own PDB file, which enables you to process unpublished data, theoretical models, or "trimmed" chains, e.g. a domain of interest from a long chain.
In brief, the ConSurf Server uses the following process by default:
- Obtains the protein sequence for the specified PDB code (or uploaded PDB file) and chain.
- Gathers closely related sequences from Swiss-Prot (or Uniprot) with a PSI-BLAST search. E value cutoff[4], number of iterations, and number of sequences to use are configurable.
- Eliminates non-unique sequences, namely, those that are 99% or more identical with another sequence.
- Does a multiple sequence alignment with MUSCLE. (Or you can upload your own MSA.)
- Constructs a phylogenetic tree. (Or you can upload your own.)
- Calculates a conservation score for each amino acid. Classifies the conservation scores into nine levels, and maps them to standard conservation level colors (see color key at the top of this page). Marks residues for which the conservation score confidence interval is too large, hence the conservation score is unreliable ("insufficient data").
- Displays the protein, colored by conservation, in interactive 3D, using FirstGlance in Jmol, Chimera, PyMOL, or Protein Explorer.
Unlike ConSurf-DB, the ConSurf Server does no filtering of the gathered sequences before constructing the MSA (except to eliminate 99% redundant sequences). If the number of sequences obtained is too small, it is up to the user to run another job with parameters adjusted to obtain more sequences. Because sequences with <99% redundancy are not filtered out, it usually takes more than the default 50 sequences to obtain an optimally informative result.
ExamplesExamples
|
At right is the pattern of evolutionary conservation and variability reported by ConSurf-DB for the alpha chain of Major Histocompatibility Complex Class I (chain A of 2vaa).
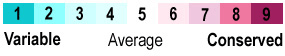
|
Because the scene at the right contains no amino acids marked insufficient data, and no chains with no data, the yellow and gray colors need not be included in the color key.
For all the available variations of the ConSurf color key, see Help:Color_Keys#ConSurf. |
2vaa contains three chains. Here, ConSurf colors are applied only to the alpha chain (chain A), while the beta chain (chain B) and the peptide (chain P) are shown as gray backbone traces. Below are instructions for how to insert a ConSurf result into a Proteopedia scene.
Examples of conserved patches on other proteins, revealed by ConSurf, will be found in the articles on
- Lac repressor
- Avian Influenza Neuraminidase, Tamiflu and Relenza
- Mechanosensitive channels: opening and closing
How to Insert a ConSurf Result Into a Proteopedia Green LinkHow to Insert a ConSurf Result Into a Proteopedia Green Link
To create a green-linked scene with a molecule colored by evolutionary conservation use the button "evolutionary conservation" in the "color" tab of the Scene Authoring Tools.
If for some reason you want to calculate the ConSurf coloring scheme on your own and want to insert that into a Proteopedia scene, here is how:
- Using either the ConSurf Database or the ConSurf Server, obtain the desired result.
- At the ConSurf result page, use the link RasMol Coloring Script to display either the script showing or hiding insufficient data. Block and copy the entire script.
- We assume that you already have an article in Proteopedia, with a Jmol applet in place for displaying your ConSurf result. (If not, see the Video Guides and Help:Editing.)
- Edit your Proteopedia page, and open the Scene Authoring Tools.
- Load the desired molecule into Jmol in the Scene Authoring Tools.
- Click on "Jmol" (at the lower right of Jmol) to open Jmol's menu, and there, click on "Console".
- In the small white Console window, paste your RasMol Coloring Script into the bottom box, and click Execute.
- Make any other changes you wish to this scene, and then save the scene.
- Copy the wikitext for the green link that will display your scene, and close the Scene Authoring Tool.
- Paste the green link wikitext into your page, and save the page.
The color key that you see at the very top of this page can be inserted in any page using this wikitext:
{{Template:ColorKey_ConSurf}}
See also Help:Color_Keys for other variations on the color key. If something is not clear, please let us know at ![]() .
.
Other Evolutionary Conservation ServersOther Evolutionary Conservation Servers
INTREPIDINTREPID
"INTREPID is an information-theoretic approach for functional site identification that exploits the information in large diverse multiple sequence alignments. INTREPID gathers homologs for a sequence using PSI-BLAST and estimates a phylogenetic tree. It then uses Jensen-Shannon divergence to measure the information for each position in the sequence at each subtree node encountered on a traversal of the phylogeny, tracing a path from the root to the leaf corresponding to the sequence of interest. Positions that are conserved across the entire family receive stronger scores than those that only become conserved within more closely related subgroups. This tree traversal produces a phylogenomic conservation score for each position in the MSA. INTREPID uses information from sequence only, and can thus be used when knowledge of structure is not available." (Quoted from the INTREPID website.)
INTREPID accepts a protein chain sequence as input. It offers to color conserved residues on 3D protein structures in Jmol. The 3D structures are obtained (when available) from the Protein Data Bank by sequence alignment searching, and users may choose from a menu of hits.
Evidence is provided that INTREPID out-performs ConSurf for predicting catalytic residues.
Unlike ConSurf, INTREPID does not identify the most variable residues in addition to the most conserved.
siteFiNDER|3DsiteFiNDER|3D
siteFiNDER|3D performs conserved functional group (CFG) analysis. "CFG Analysis is a general method for predicting the location of functionally important sites within a target protein structure. Like other available structure/sequence analysis techniques, CFG Analysis exploits the evolutionary relationships present across groups of homologous proteins to identify regions that are likely to be of functional significance. However, this technique is particularly useful for situations where other methods fail, for instance when only a few or highly similar homologues can be identified." As its name implies, CFG analysis attempts to identify groups of conserved amino acids that together represent a functional site. In this respect, it goes beyond most other evolutionary conservation servers, which stop at assigning a conservation value to each amino acid. See the comparison of siteFiNDER|3D with ConSurf for cytochrome c.
This site provides links to several other software packages that predict functional sites, some of which are not further discussed in the present article.
HotPatchHotPatch
HotPatch [11] "finds unusual patches on the surface of proteins, and computes just how unusual they are (patch rareness), and how likely each patch is to be of functional importance (functional confidence (FC).) The statistical analysis is done by comparing your protein's surface against the surfaces of a large set of proteins whose functional sites are known." One advantage of HotPatch is that sequence homologs are not required. See the comparison of HotPatch with ConSurf for cytochrome c.
Evolutionary Trace ViewerEvolutionary Trace Viewer
Evolutionary Trace Viewer (ETV). See the comparison of ETV with ConSurf for cytochrome c.
Comment by User:Eric Martz, March, 2009: From the information provided on the ETV website, I found it quite difficult to understand what the ETV is doing, or how to use the viewer. An explanation in simple terms for non-specialists would be very useful.
NotesNotes
- ↑ 1.0 1.1 1.2 1.3 Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009 Jan;37(Database issue):D323-7. Epub 2008 Oct 29. PMID:18971256 doi:http://dx.doi.org/10.1093/nar/gkn822
- ↑ PSI-BLAST (Position Specific Iteration-BLAST) is an extension of the Basic Local Alignment Search Tool (BLAST) that is more sensitive at finding distantly related sequences. See PSI-BLAST at Wikipedia and PSI-BLAST at NCBI.
- ↑ From UniProtKB help: "UniProtKB/Swiss-Prot (reviewed) is a high quality manually annotated and non-redundant protein sequence database, which brings together experimental results, computed features and scientific conclusions."
- ↑ 4.0 4.1 4.2 4.3 Expectation Value (E value): When searching a sequence database with a query sequence, e.g. using BLAST or PSI-BLAST, each found sequence can be characterized by an E value. It is the number of hits expected by chance with the sequence matching level observed, taking into account the size of the sequence database and length of the query sequence. Low values of E (much less than one) mean increasing significance of the match.
- ↑ Mayrose I, Graur D, Ben-Tal N, Pupko T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol Biol Evol. 2004 Sep;21(9):1781-91. Epub 2004 Jun 16. PMID:15201400 doi:http://dx.doi.org/10.1093/molbev/msh194
- ↑ Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992 Jun;8(3):275-82. PMID:1633570
- ↑ Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006 Jul 1;22(13):1658-9. Epub 2006 May 26. PMID:16731699 doi:http://dx.doi.org/10.1093/bioinformatics/btl158
- ↑ Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001 Mar 16;307(1):447-63. PMID:11243830 doi:http://dx.doi.org/10.1006/jmbi.2000.4474
- ↑ Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003 Jan;19(1):163-4. PMID:12499312
- ↑ Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W299-302. PMID:15980475 doi:http://dx.doi.org/33/suppl_2/W299
- ↑ Pettit FK, Bare E, Tsai A, Bowie JU. HotPatch: a statistical approach to finding biologically relevant features on protein surfaces. J Mol Biol. 2007 Jun 8;369(3):863-79. Epub 2007 Mar 21. PMID:17451744 doi:http://dx.doi.org/10.1016/j.jmb.2007.03.036