User:Eric Martz/Sandbox 6: Difference between revisions
Eric Martz (talk | contribs) |
Eric Martz (talk | contribs) |
||
| Line 87: | Line 87: | ||
<scene name='User:Eric_Martz/Sandbox_6/Adafaf/2'>TextToBeDisplayed</scene> | <scene name='User:Eric_Martz/Sandbox_6/Adafaf/2'>TextToBeDisplayed</scene> | ||
<scene name='User:Eric_Martz/Sandbox_6/3ckz_relenza_tyr274/2'>TextToBeDisplayed</scene> | |||
Revision as of 04:00, 30 April 2009
Avian Influenza Neuraminidase, Tamiflu and Relenza
Influenza Virus NeuraminidaseInfluenza Virus Neuraminidase
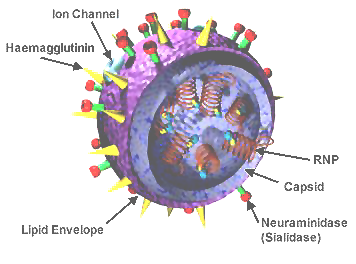
The surfaces of influenza viruses include, among other molecules, two glycoproteins named hemagglutinin and neuraminidase, coded for by the viral segmented RNA genome. Each of these molecules is required for successful infection and spread in a host animal. The hemagglutinin attaches influenza to sialic acid on the surfaces of cells, enabling them to enter and infect cells. After the virus has replicated, neuraminidase (also called sialidase) removes sialic acid from the cell, enabling the newly assembled virions to be released in order to spread and infect other cells. For more about the structure and biology, including references for the points made here, please see Influenza at Wikipedia.
|
- Influenza neuraminidase is a homotetramer[2] ().
- Each of the four protein chains in the tetramer has a catalytic site, indicated in by the positions of the bound Tamiflu inhibitors.
- The involves only a single protein chain, being distant from neighboring chains.
- The is mostly beta, consisting of several beta sheets with three short alpha helices ( Beta Strands , Alpha Helices).
- The residues contacting the Tamiflu inhibitory substrate analog are [3].

- These highly conserved residues include some known to be crucial to binding sialic acid substrate: Arg 118, Arg 292 and Arg 371 bind the carboxylate; Arg 152 interacts with the acetamido substituent; and Glu 276 forms hydrogen bonds with the 8- and 9-hydroxyl groups of the substrate. These residues are in contact with the sialic acid substrate analog Tamiflu. In this scene, atoms and bonds in Tamiflu and the highlighted residues are colored by element: C, O, N.
Past Influenza PandemicsPast Influenza Pandemics
Influenza A (one of several genera and species of influenza) is the most virulent form infecting humans, killing 500,000 people worldwide annually (including about 36,000 in the USA). Its hemagglutinin (H) and neuraminidase (N) are classified into various numbered serotypes or subtypes, such as H1N1, H2N2, H3N2, H5N1, and so forth[4].
- H1N1 1918-20: The Spanish Flu pandemic killed tens of millions of people worldwide (about twice as many as were killed in World War I). One in five suffered with this disease, and about one in 30 died. This pandemic may have killed more people than did the Black Plague. Spanish Flu was caused by a particularly virulent form of H1N1 believed to be derived from influenza A viruses in the natural reservoir of wild birds.
- H2N2 1957-58: The Asian Flu pandemic originated when a virus mutation in the wild duck reservoir of influenza virus combined with a human strain. This virus, of subtype H2N2, killed nearly 70,000 people in the USA and infected millions worldwide. It was contained in part by a vaccine developed during the pandemic. H2N2 is also suspected of causing the Russian Flu pandemic that killed about one million people in 1889-90.
- H3N2 1968-69: The Hong Kong Flu pandemic was caused by an H3N2 subtype derived by genetic recombination (antigenic shift) between virus subtypes, believed to have occurred during co-infection of pigs by multiple virus subtypes including H2N2. Although it infected hundreds of millions of people worldwide, its modest virulence prevented the death rate from greatly exceeding that of normal flu seasons.
H5N1 Pandemic ThreatH5N1 Pandemic Threat
A new influenza pandemic is one of our greatest threats because it might well kill a large fraction of the human population. The subtype H5N1 is most feared because of the large reservoir in wild birds, and the recent emergence of strains called highly pathogenic avian influenza (HPAI) that have high virulence and mortality in birds. Hundreds of millions of domestic poultry have been culled at great economic cost in an effort to stem the spread of H5N1. Although transmission from birds to humans has apparently been very inefficient, over half of the people known to have been infected died from the disease. The emergence of a high-virulence form of H5N1 that is highly transmissable among humans seems nearly inevitable, and would cause a devastating pandemic.
Prophylaxis and Treatment of InfluenzaProphylaxis and Treatment of Influenza
Vaccines are effective at preventing influenza, but only if they target the relevant viral subtypes. New vaccines against the annual epidemics of influenza A and B are prepared each year, separately in the northern and southern hemispheres. These are designed to target the subtypes predicted to be prevalent in any given flu season, but sometimes those predictions are wrong, leading to that year's vaccine being ineffective. A vaccine for a pandemic strain of H5N1 could not be prepared until after the pandemic began, because only then would the relevant subtype be known[5].
Drugs against influenza, stockpiled in advance of a panedmic, appear to be the best preparation, given the limitations of vaccines. Tens of billions of dollars have been spent on pandemic preparedness in the USA alone, and a large portion of these expenditures is for stockpiling of anti-influenza drugs. Similar expenditures have been made in many developed countries. The World Health Organization is poised to distribute anti-influenza drugs at the first signs of an epidemic of H5N1.
Amantadine and RimantadineAmantadine and Rimantadine
Amantadine and Rimantadine are an anti-viral drugs believed to work by blocking an ion-channel (M2) required for viruses to infect cells. Ion-channel function appears to be required for uncoating during endocytosis. Amantadine was approved for anti-viral uses beginning in 1966 by the US FDA. Subsequent widespread use has selected amantadine-resistant influenza in humans and birds. By 2005-2006, the US CDC found 92% of H3N2 isolates were resistant, and 2 of 8 H1N1 isolates. In Asia, resistance is close to 100%. The most common mutation responsible for resistance is S31N in M2, which confers resistance to both amantadine and rimantadine[6]. References for this paragraph will be found in the Amantadine article in Wikipedia.
Tamiflu® (oseltamivir) and Relenza® (zanamivir)Tamiflu® (oseltamivir) and Relenza® (zanamivir)
Tamiflu (oseltamivir) is an inhibitor of influenza neuraminidase that binds to the enzyme active site. (Tamiflu is Roche's trade name; oseltamivir is the generic name.) Tamiflu is a transition state analog, and was the first orally active neuraminidase inhibitor commercially developed. Because neuraminidase is required for the viral life cycle, its enzymatic active site is highly conserved, and Tamflu is effective on a range of neuraminidase subtypes. It is indicated both for prophylaxis and for treatment within two days of the onset of symptoms.
Relenza (zanamivir) is also an inhibitor of influenze neuraminidase that binds to the enzyme active site. (Relenza is GlaxoSmithKline's trade name; zanamivir is the generic name.) Unlike Tamiflu, which is given orally, Relenza is usually administered by inhalation, or can be injected.
Structure-based drug design was employed in the development of both Tamiflu and Relenza[7] . A structure of N2 at 2.9 Å resolution was published in 1983[8], and a 2.2 Å structure, 1nn2, was deposited by the same authors in the PDB in 1991. The structure of N9 was determined by the same group, e.g. 7nn9.
Because Tamiflu and Relenza closely resemble the natural sialic acid substrate of neuraminidase, it was hoped that mutations conferring resistance to these drugs would greatly lower the virulence of influenza carrying such mutations. This hope has proven false in the case of Tamiflu[9]. However, two common mutations that confer resistance to Tamiflu did not confer resistance to Relenza[9]. This suggests that it would be prudent to stockpile Relenza in addition to Tamiflu, and that combination therapy might be the most effective weapon against a new pandemic, prior to development and deployment of a vaccine.
Tamiflu Binds to N1 by Induced FitTamiflu Binds to N1 by Induced Fit
|
Tamiflu was designed to fit N2/N9, so it is serendipitous that it works on N1. In fact, when the structure of N1 was determined[7], the loop comprising residues 147-152 was not in a suitable position to participate in binding Tamiflu. However, the complex of N1 with Tamiflu revealed that this loop is pulled into proper contact with the drug in an induced fit manner[7]. A morph from N1 alone (2hty) to N1 complexed with Tamiflu (2hu4)[10] shows the change in position of this loop ().
The binding of Tamiflu to N1 pulls the sidechains of two conserved residues, , closer to the inhibitor.
Cavity in N1: An Opportunity for Drug DesignCavity in N1: An Opportunity for Drug Design
N1 has a larger surface-accessible cavity in the substrate binding region than is present in N2/N9. The larger end of this cavity is not occupied by Tamiflu. Thus, this cavity presents an opportunity to design a drug with greater specificity and potency for N1[7].
For technical reasons, the cavity cannot be shown yet in Jmol in Proteopedia. (We are working to resolve this problem.) However, it may be seen in View 2 of this Chapter of the Jmol Tutorial-Authoring Template (JTAT) Demonstration Tutorial.
Tamiflu-Resistant MutantsTamiflu-Resistant Mutants
|
LinksLinks
- cdc.gov/flu, the official influenza resource of the US Center for Disease Control.
- Pandemic Planning Toolkit (by Roche).
- Relenza offical website by GlaxoSmithKline.
- Tamiflu official website by Roche.
- The Next Pandemic? An authoritative overview of economic and political factors written in 2005 by Laurie Garrett.
Notes and Literature ReferencesNotes and Literature References
- ↑ Image of influenza virus structure was obtained from Wikipedia.
- ↑ The tetramer is one of two biological units in the asymmetric unit of 2hu4.
- ↑ See Evolutionary Conservation. Coloring by ConSurf on chain A of 2hu4 based on 100 unique homologs using default conditions, done on September 23, 2008.
- ↑ Influenza at Wikipedia.
- ↑ Vaccination for Influenza at Wikipedia
- ↑ Report on amantadine resistance, CDC Morbidity and Mortality Weekly Reports, January 2006.
- ↑ 7.0 7.1 7.2 7.3 The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design, Russell et al., Nature 443:45, 2006. PubMed 16915235.
- ↑ Varghese, Laver & Colman, Nature 303:35, 1983. PubMed 6843658
- ↑ 9.0 9.1 Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants, Collins et al., Nature 453:1248, 2008. PubMed 18480754.
- ↑ Chain A from 2hty was morphed to chain A of 2hu4 by linear interpolation, inserting 6 intermediate interpolated frames, using the freely available morph2 program.