Rebecca Martin/Sandbox1: Difference between revisions
No edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
== Antibody Structure and the Immunoglobulin Domain == | == Antibody Structure and the Immunoglobulin Domain == | ||
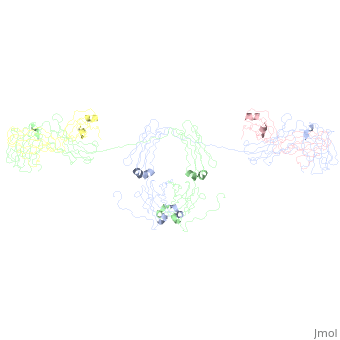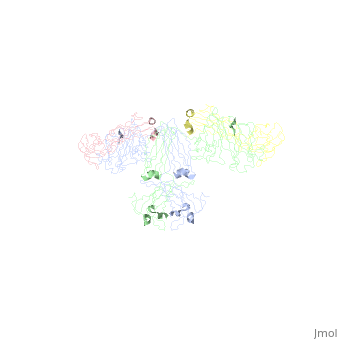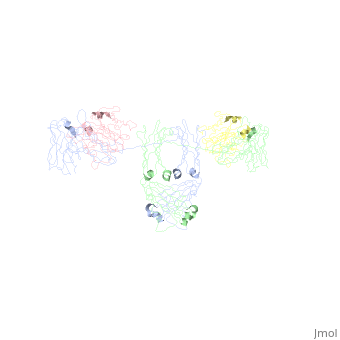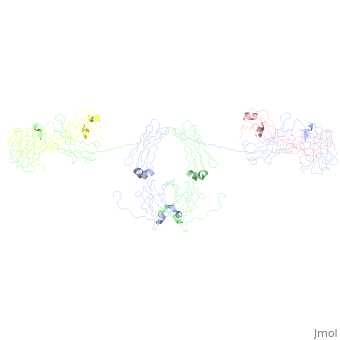
An antibody is a tetramer of 2 <scene name='Rebecca_Martin/Sandbox1/ | An antibody is a tetramer of 2 <scene name='Rebecca_Martin/Sandbox1/Iga1_light_chains/1'>light_chains</scene> and 2 <scene name='Rebecca_Martin/Sandbox1/Iga1_heavy_chains/1'>heavy chains</scene>. In other words, the antibody is a homodimer of 2 heterodimers. Each <scene name='Rebecca_Martin/Sandbox1/Iga1_homodimer/1'>heterodimer</scene> is comprised on one light chain and one heavy chain. Heavy and light chains are held together with disulfide bonds and noncovalent interactions (more on this later). | ||
Another common way of describing antibody structure is | Another common way of describing antibody structure is | ||
Revision as of 10:30, 23 April 2009
Introduction to IgAIntroduction to IgA
Template:STRUCTURE 1iga The most extensive surface in contact with the external environment is not our skin, but the epithelial lining of our gastrointestinal, respiratory, and urogenital tracts [1]. As a first line of defense in maintainance the integrity our mucosa, the immune system manufatures and secretes dimeric IgA to neutralize pathogenic organisms [2] and exclude the entry of commensals at the mucosal border [3]. In the serum, IgA functions as a second line of defense against pathogens that may breech the epithelial boundary [2]. The body produces more IgA than any other antibody isotype [3]. In fact, is the most abundant antibody in the body, further illustrating IgA's critical role in immunity [4].
Unlike other antibody isotypes, IgA exists in mutiple oligomeric states [3]. The most common of which are the monomeric, dimeric, and secretory forms [4]. At least two isotypes exist, termed IgA1 and IgA2. IgA2 can further be categorized into 2 allotypes: IgA2 m(1) and IgA2 m(2). The receptors for IgA include the Fcα Receptor (FcαRI; CD89) and the polyimmunologlobulin receptor (pIgRI). When binding to FcαRI results in the dimerization, the consequent signaling results in effector functions, including respiratory burstmucosal surface, an approximately equal ratio of secretory IgA1 (sIgA1) to secretory IgA2 (sIgA2) reside at the mucosal surface, with the exception of the colon, where the majority is sIgA2 [5]. In the serum, about 90% of the IgA is monomeric IgA1 [4], phaocytosis, and eosinophil degranulation. Binding to the pIgR results in transoocytosis and IgA secretion [2]. Exploring IgA's structure and protein interactions illuminates the unique and critical function IgA plays in humoral immunity.
Antibody Structure and the Immunoglobulin DomainAntibody Structure and the Immunoglobulin Domain
An antibody is a tetramer of 2 and 2 . In other words, the antibody is a homodimer of 2 heterodimers. Each is comprised on one light chain and one heavy chain. Heavy and light chains are held together with disulfide bonds and noncovalent interactions (more on this later).
Another common way of describing antibody structure is
Each light chains are composed of 2 immunoglobulin domains: one and one constant domain. Heavy chains composed of 4 Ig domains: one V-type and 3 C-type, named CH1 - CH3. A linking hinge region separates the CH2 and CH3 domains. This site is subject to proteolytic cleavage by the protease papain, which fragments the antibody into Fab and Fc fragments.
The antibody is a member of the immunoglobulin superfamily of proteins (ref Att), and can be further broken down into 12 immunoglobulin domains. Each immunoglobulin domain contains a primary amino acid sequence of approximately 70 – 100 residues long. Secondary structure is a characteristic beta sandwich with a variable number of beta strands, depending on the unit type. These strands reside in a Greek Key connectivity (web other) and form 2 beta sheets that fold over each other. A disulfide bond stabilizes the tertiary structure.
Nine antiparrallel beta strands comprise variable or V-regions. Loop sequences of varying length connect the strands. The 9 strands form 2 beta sheets, one with 4 (ABED-prosite) strands and the other with 3 (CFG prosite). The remaining 2 strands (C’ and C”) lie in between the 2 sheets. A disulfide bride stabilizes the 2 sandwich halves. Hydrophobic residues face the interior of the sheet, providing stability, while hydrophillic residues face outward and interact with the local environment. The extra loops in the V-region is critical for epitope specificity, and are consequently known as the compliment determining regions.
C-type domains lack the 2 sandwiched beta strands. The sheets are ABED and CFG. Consequently, the sandich is more tightly packed.
While the classic Ig like domain is found predominantly in the immune system and represents the closest related structires: The most closely related molecules are the antigen recognition molecules: MHC, Igs, TCRs are the MHC receptor
The V-type comprises a wide variety including the Ig binding molecules including the pIgR and the FcalphaR. The SC = 4 Vtype domains, more cloc=sely related to eachother – duplication events.
Viral hemagluttinin is another example.
Variable domains determine antigen specificity. The loops @ ___ are the most variable regions, and are known as compliment determining regions
Constant regions determine the isotype: IgA, IgD, IgM, IgG, or IgE
To
- Immunoglobulin Structure
Antibodies are composed of a heavy chain and a light chain.
Fab fragment
- Forms of IgA
Dimeric Structure
|
- Secretory Component
Monomeric IgAMonomeric IgA
Monomeric iga: 12 domains ~ IgG 10064707 2 light, 2 heavy. 10064707 2 domains in Light = vl and cl. 10064707 4 domains in heavy. Heavy = vh, c1, c2, c3 10064707 Gen structure of domains = beta sheet sandwhich structure 10064707 + a conserved disulphide bond 15111057 Beta strands = A to G 10064707 V domains = 9 strands (DEBA-GFCC’C’’) 10064707 C domains = 7 or 8 strands DEBA-GFC(C’) 10064707 Disulfide Cys 311-Cys 471 10064707 90% monomeric IgA1) 10064707
|
|
|
|
Dimeric IgA and the J chainDimeric IgA and the J chain
Secretory IgA and the Secretory ComponentSecretory IgA and the Secretory Component
|
Insights into FunctionInsights into Function
Implications in Science and MedicineImplications in Science and Medicine
Evolution and Related LinksEvolution and Related Links
Limitations of the Current StudiesLimitations of the Current Studies
- 10064707; 15111057 xray and neutron scattering analysis + analytical ultracentrifugation and analyzed w constrained modeling 2/2 high carb and flex = difficult to crystalize 18178841
Questions for the FutureQuestions for the Future
- Because of the limitating resolution of these models, many details concerning the binding residues and residue interactions are left unknown. Crystallographic structure will yield further insights into the structure of IgA, the interactions between IgA and other molecules, and ....
SC aa interact w J chain? CDR-like motifs @ D1 binds ? @ IgA; Does SC open upon binding?; stoichiometry of binding? Locations of oligos on SC? Differences in binding IgA1 vs IgA2 17428798 Binding motifs SC and IgA1 18178841 Structure of IgA involved in IgA nephropathy 18178841
LinksLinks
IgAIgA
- Fab and Fc Fragments
- Refined crystal structure of the galactan-binding immunoglobulin fab j539 at 1.95-angstroms resolution 2fbj
- Phosphocholine binding immunoglobulin fab mc/pc603. an x-ray diffraction study at 2.7 angstroms 1mcp
- Phosphocholine binding immunoglobulin fab mc/pc603. an x-ray diffraction study at 3.1 angstroms 2mcp
- Crystal structure of human FcaRI bound to IgA1-Fc 1ow0
- Refined crystal structure of a recombinant immunoglobulin domain and a complementarity-determining region 1-grafted mutant 2imm and2imn
- Crystal structure of a Staphylococcus aureus protein (SSL7) in complex with Fc of human IgA1 2qej
- Monomeric
- Dimeric and Secretory
Related MoleculesRelated Molecules
- non-IgA antibody isotypes
- IgM: Solution structure of human Immunoglobulin M 2rcj
- IgG:
- IgD:
- IgE:
- Other C-type immunoglobulin examples
- MHC
- TCR
- V-type immunoglobulin examples
ReferencesReferences
- ↑ Bonner A, Perrier C, Corthesy B, Perkins SJ. Solution structure of human secretory component and implications for biological function. J Biol Chem. 2007 Jun 8;282(23):16969-80. Epub 2007 Apr 11. PMID:17428798 doi:http://dx.doi.org/10.1074/jbc.M701281200
- ↑ 2.0 2.1 2.2 Furtado PB, Whitty PW, Robertson A, Eaton JT, Almogren A, Kerr MA, Woof JM, Perkins SJ. Solution structure determination of monomeric human IgA2 by X-ray and neutron scattering, analytical ultracentrifugation and constrained modelling: a comparison with monomeric human IgA1. J Mol Biol. 2004 May 14;338(5):921-41. PMID:15111057 doi:http://dx.doi.org/10.1016/j.jmb.2004.03.007
- ↑ 3.0 3.1 3.2 Bonner A, Almogren A, Furtado PB, Kerr MA, Perkins SJ. Location of secretory component on the Fc edge of dimeric IgA1 reveals insight into the role of secretory IgA1 in mucosal immunity. Mucosal Immunol. 2009 Jan;2(1):74-84. Epub 2008 Oct 8. PMID:19079336 doi:http://dx.doi.org/10.1038/mi.2008.68
- ↑ 4.0 4.1 4.2 Boehm MK, Woof JM, Kerr MA, Perkins SJ. The Fab and Fc fragments of IgA1 exhibit a different arrangement from that in IgG: a study by X-ray and neutron solution scattering and homology modelling. J Mol Biol. 1999 Mar 12;286(5):1421-47. PMID:10064707 doi:http://dx.doi.org/10.1006/jmbi.1998.2556
- ↑ Bonner A, Almogren A, Furtado PB, Kerr MA, Perkins SJ. The nonplanar secretory IgA2 and near planar secretory IgA1 solution structures rationalize their different mucosal immune responses. J Biol Chem. 2009 Feb 20;284(8):5077-87. Epub 2008 Dec 23. PMID:19109255 doi:http://dx.doi.org/10.1074/jbc.M807529200