2cts: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
<StructureSection load='2cts' size='340' side='right'caption='[[2cts]], [[Resolution|resolution]] 2.00Å' scene=''> | <StructureSection load='2cts' size='340' side='right'caption='[[2cts]], [[Resolution|resolution]] 2.00Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
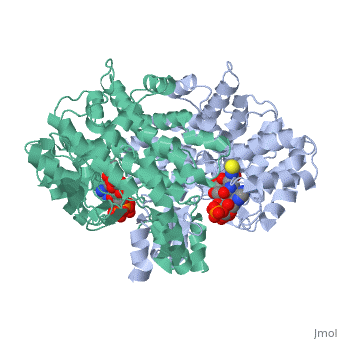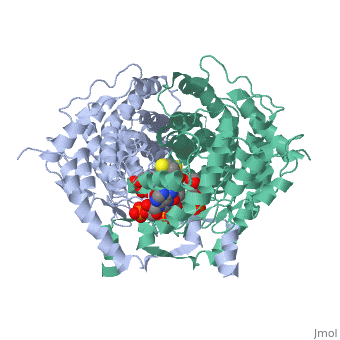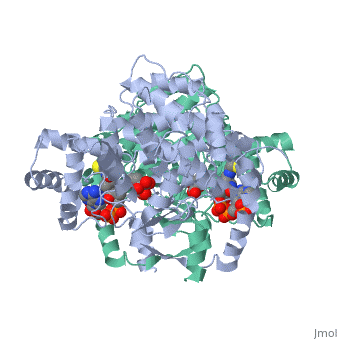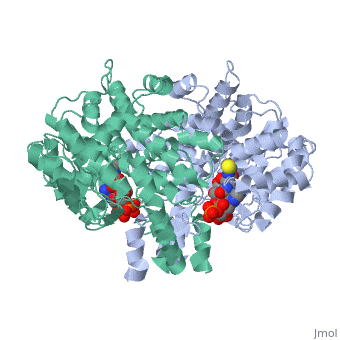
<table><tr><td colspan='2'>[[2cts]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/ | <table><tr><td colspan='2'>[[2cts]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Sus_scrofa Sus scrofa]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2CTS OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2CTS FirstGlance]. <br> | ||
</td></tr><tr id=' | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2Å</td></tr> | ||
<tr id=' | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CIT:CITRIC+ACID'>CIT</scene>, <scene name='pdbligand=COA:COENZYME+A'>COA</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2cts FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2cts OCA], [https://pdbe.org/2cts PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2cts RCSB], [https://www.ebi.ac.uk/pdbsum/2cts PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2cts ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2cts FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2cts OCA], [https://pdbe.org/2cts PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2cts RCSB], [https://www.ebi.ac.uk/pdbsum/2cts PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2cts ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | |||
[https://www.uniprot.org/uniprot/CISY_PIG CISY_PIG] | |||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 25: | Line 27: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
[[Category: | [[Category: Sus scrofa]] | ||
[[Category: Huber | [[Category: Huber R]] | ||
[[Category: Remington | [[Category: Remington S]] | ||
[[Category: Wiegand | [[Category: Wiegand G]] | ||
Latest revision as of 12:18, 14 February 2024
CRYSTALLOGRAPHIC REFINEMENT AND ATOMIC MODELS OF TWO DIFFERENT FORMS OF CITRATE SYNTHASE AT 2.7 AND 1.7 ANGSTROMS RESOLUTIONCRYSTALLOGRAPHIC REFINEMENT AND ATOMIC MODELS OF TWO DIFFERENT FORMS OF CITRATE SYNTHASE AT 2.7 AND 1.7 ANGSTROMS RESOLUTION
Structural highlights
FunctionEvolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. See Also |
| ||||||||||||||||||