2af5: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
<StructureSection load='2af5' size='340' side='right'caption='[[2af5]], [[Resolution|resolution]] 2.50Å' scene=''> | <StructureSection load='2af5' size='340' side='right'caption='[[2af5]], [[Resolution|resolution]] 2.50Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[2af5]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/ | <table><tr><td colspan='2'>[[2af5]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Borreliella_burgdorferi Borreliella burgdorferi]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2AF5 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2AF5 FirstGlance]. <br> | ||
</td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2af5 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2af5 OCA], [https://pdbe.org/2af5 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2af5 RCSB], [https://www.ebi.ac.uk/pdbsum/2af5 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2af5 ProSAT]</span></td></tr> | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2af5 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2af5 OCA], [https://pdbe.org/2af5 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2af5 RCSB], [https://www.ebi.ac.uk/pdbsum/2af5 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2af5 ProSAT]</span></td></tr> | |||
</table> | </table> | ||
== Function == | |||
[https://www.uniprot.org/uniprot/OSPA_BORBU OSPA_BORBU] | |||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 32: | Line 35: | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
[[Category: | [[Category: Borreliella burgdorferi]] | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
[[Category: Hilyard | [[Category: Hilyard A]] | ||
[[Category: Koide | [[Category: Koide A]] | ||
[[Category: Koide | [[Category: Koide S]] | ||
[[Category: Makabe | [[Category: Makabe K]] | ||
[[Category: Mcelheny | [[Category: Mcelheny D]] | ||
[[Category: Tereshko | [[Category: Tereshko V]] | ||
Latest revision as of 10:23, 23 August 2023
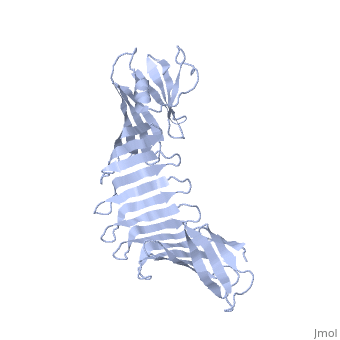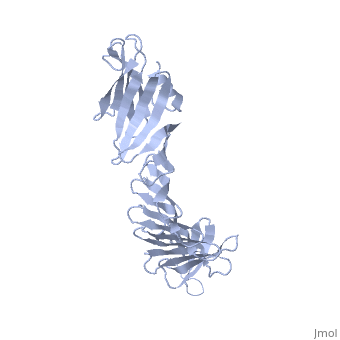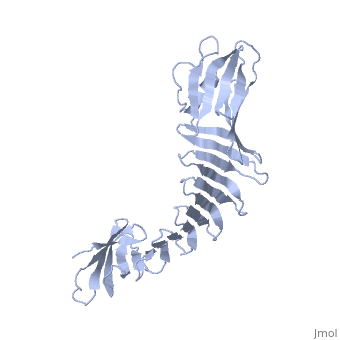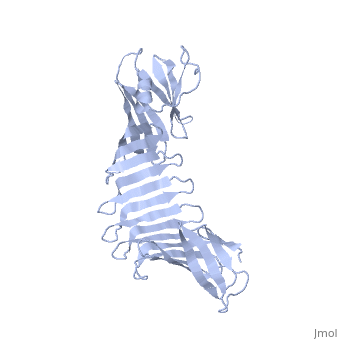
2.5A X-ray Structure of Engineered OspA protein2.5A X-ray Structure of Engineered OspA protein
Structural highlights
FunctionEvolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedAlthough the beta-rich self-assemblies are a major structural class for polypeptides and the focus of intense research, little is known about their atomic structures and dynamics due to their insoluble and noncrystalline nature. We developed a protein engineering strategy that captures a self-assembly segment in a water-soluble molecule. A predefined number of self-assembling peptide units are linked, and the beta-sheet ends are capped to prevent aggregation, which yields a mono-dispersed soluble protein. We tested this strategy by using Borrelia outer surface protein (OspA) whose single-layer beta-sheet located between two globular domains consists of two beta-hairpin units and thus can be considered as a prototype of self-assembly. We constructed self-assembly mimics of different sizes and determined their atomic structures using x-ray crystallography and NMR spectroscopy. Highly regular beta-sheet geometries were maintained in these structures, and peptide units had a nearly identical conformation, supporting the concept that a peptide in the regular beta-geometry is primed for self-assembly. However, we found small but significant differences in the relative orientation between adjacent peptide units in terms of beta-sheet twist and bend, suggesting their inherent flexibility. Modeling shows how this conformational diversity, when propagated over a large number of peptide units, can lead to a substantial degree of nanoscale polymorphism of self-assemblies. Atomic structures of peptide self-assembly mimics.,Makabe K, McElheny D, Tereshko V, Hilyard A, Gawlak G, Yan S, Koide A, Koide S Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17753-8. Epub 2006 Nov 8. PMID:17093048[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences |
| ||||||||||||||||