Alternate locations of backbones: Difference between revisions
Eric Martz (talk | contribs) |
Eric Martz (talk | contribs) |
||
| Line 113: | Line 113: | ||
==AltLoc Backbone Cases== | ==AltLoc Backbone Cases== | ||
====Case 1: Flexible active site loop broadens range of protection.==== | |||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
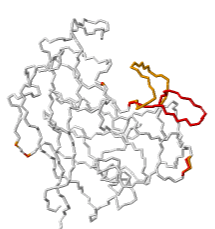
| | | [https://www.uniprot.org/uniprotkb/B0LJC8/entry TRIMCyp protein] mediates innate immunity to HIV by inhibiting post-entry retrovirus replication. Evidence suggests that TRIMCyp has evolved through mutations to broaden the range of retroviruses it can inhibit<ref name="4dgd">PMID: 22407016</ref>. These mutations render the active site able to assume multiple conformations. In particular, the cyclophilin-binding loop 64-74 (GGNFT HCNGT GG), flanked by double glycine pivots, exhibited flexibility<ref name="4dgd" />. A crystal structure, [[4dgd]], resolved two alternate location conformations for the loop residues 66-74, shown at right in red (AltLoc A) and orange (AltLoc B), each at about 50% occupancy. | ||
Explore [http://firstglance.jmol.org/fg.htm?mol=4dgd 4DGD in FirstGlance in Jmol], which has [[Alternate_locations#Visualizing_alternate_locations|AltLoc visualization/animation capabilities]]. | Explore [http://firstglance.jmol.org/fg.htm?mol=4dgd 4DGD in FirstGlance in Jmol], which has [[Alternate_locations#Visualizing_alternate_locations|AltLoc visualization/animation capabilities]]. | ||
| Line 123: | Line 124: | ||
|} | |} | ||
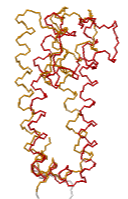
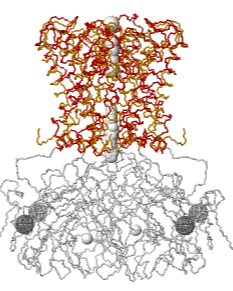
====Case 2: Rotation of potassium channel regulates gating.==== | |||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| rowspan=2 | | | rowspan=2 | mmm | ||
Explore [http://firstglance.jmol.org/fg.htm?mol=2wlk 2WLK in FirstGlance in Jmol], which has [[Alternate_locations#Visualizing_alternate_locations|AltLoc visualization/animation capabilities]]. | Explore [http://firstglance.jmol.org/fg.htm?mol=2wlk 2WLK in FirstGlance in Jmol], which has [[Alternate_locations#Visualizing_alternate_locations|AltLoc visualization/animation capabilities]]. | ||
Revision as of 21:07, 8 June 2023
At sufficiently high resolution, empirical methods for determining macromolecular structure may detect multiple locations for some atoms, termed alternate locations. When alternate locations for backbones (main chains) deviate substantially, the backbone may separate into two pathways ("fork") and then rejoin into a single pathway.
Alternate location serverAlternate location server
An Alternate Location Server (AltLoc Server) (part of the OCA Browser system) scans all entries in the Protein Data Bank, listing those that have alternate locations. The analysis on this page is based on an AltLoc Server report from late May, 2023, when the total number of entries in the PDB was ~200,700. Here are two example lines from the report:
ID Method Res Dep Date 9ins X-ray diffraction 1.70 1991-10-23 min=-1.0 ave=-1.0 max=-1.0 A=48 B=48 4dgd X-ray diffraction 1.4 2012-01-25 min=0.03 ave=4.78 max=15.87 A=84 B=84 A.CA=13 B.CA=13
- Res is the resolution in Å.
- Dep Date is the date of deposition into the PDB.
- min, ave, max are for the distances between AltLocs of backbone atoms only. "Backbone atoms" are protein alpha carbon atoms, and nucleic acid phosphorus atoms. A value of -1 is given when no backbone atoms have AltLocs.
- A and B are the AltLoc code characters used in these cases. Any alphanumeric characters are legal, and atoms may have more than two AltLocs.
- The numbers after A= and B= are the numbers of atoms with A/B AltLoc codes.
- The numbers after A.CA= and B.CA= are the numbers of alpha carbon atoms with A/B AltLoc codes. For 9ins, the absence of .CA= counts means that no alpha carbons have AltLocs in this entry.
Prevalence of AltLocsPrevalence of AltLocs
The prevalence of AltLocs is summarized in the article Alternate locations.
Backbone Altloc SeparationsBackbone Altloc Separations
X-ray diffractionX-ray diffraction
| Range of Separation Averages, Å | Entry Count | Average Resolution, Å |
|---|---|---|
| 0.00-0.20 | 70,134 | 1.65 |
| 0.21-0.50 | 4,446 | 1.64 |
| 0.51-1.00 | 1,358 | 1.66 |
| 1.01-2.50 | 1,128 | 1.86 |
| 2.51-5.00 | 307 | 2.08 |
| 5.01-10.00 | 99 | 2.23 |
| 10.01-20.00 | 39 | 2.16 |
| 20.01-40.00 | 40 | 2.26 |
| ≥ 40.01 | 6 | 2.62 |
Electron MicroscopyElectron Microscopy
| Range of Separation Averages, Å | Entry Count | Average Resolution, Å |
|---|---|---|
| 0.00-0.20 | 1,093 | 3.76 |
| 0.21-0.50 | 33 | 7.13 |
| 0.51-1.00 | 23 | 4.65 |
| 1.01-2.50 | 25 | 3.90 |
| 2.51-5.00 | 9 | 5.42 |
| 5.01-10.00 | 11 | 3.47 |
| 10.01-20.00 | 2 | 8.65 |
| 20.01-40.00 | 2 | 3.71 |
| ≥ 40.01 | 0 |
NMRNMR
Structures determined by NMR typically represent alternate locations or flexibility as multiple models. Only a handful of the ~14,000 NMR entries (May, 2023) in the PDB have AltLoc codes, and these are not used in the conventional manner.
AltLoc Backbone CasesAltLoc Backbone Cases
Case 1: Flexible active site loop broadens range of protection.Case 1: Flexible active site loop broadens range of protection.
| TRIMCyp protein mediates innate immunity to HIV by inhibiting post-entry retrovirus replication. Evidence suggests that TRIMCyp has evolved through mutations to broaden the range of retroviruses it can inhibit[1]. These mutations render the active site able to assume multiple conformations. In particular, the cyclophilin-binding loop 64-74 (GGNFT HCNGT GG), flanked by double glycine pivots, exhibited flexibility[1]. A crystal structure, 4dgd, resolved two alternate location conformations for the loop residues 66-74, shown at right in red (AltLoc A) and orange (AltLoc B), each at about 50% occupancy.
Explore 4DGD in FirstGlance in Jmol, which has AltLoc visualization/animation capabilities. 4dgd X-ray diffraction 1.4 2012-01-25 min=0.03 ave=4.78 max=15.87 A=84 B=84 A.CA=13 B.CA=13 |
 |
Case 2: Rotation of potassium channel regulates gating.Case 2: Rotation of potassium channel regulates gating.
| mmm
Explore 2WLK in FirstGlance in Jmol, which has AltLoc visualization/animation capabilities. 2wlk X-ray diffraction 2.8 2009-06-24 min=1.04 ave 5.64 max 9.99 A=1500 B=1540 A.CA=202 B.CA=206 |
|
|
| xxx | yyy |
ReferencesReferences
- ↑ 1.0 1.1 Caines ME, Bichel K, Price AJ, McEwan WA, Towers GJ, Willett BJ, Freund SM, James LC. Diverse HIV viruses are targeted by a conformationally dynamic antiviral. Nat Struct Mol Biol. 2012 Mar 11. doi: 10.1038/nsmb.2253. PMID:22407016 doi:10.1038/nsmb.2253