2hh7: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
<StructureSection load='2hh7' size='340' side='right'caption='[[2hh7]], [[Resolution|resolution]] 2.55Å' scene=''> | <StructureSection load='2hh7' size='340' side='right'caption='[[2hh7]], [[Resolution|resolution]] 2.55Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[2hh7]] is a 1 chain structure with sequence from [ | <table><tr><td colspan='2'>[[2hh7]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/"bacillus_tuberculosis"_(zopf_1883)_klein_1884 "bacillus tuberculosis" (zopf 1883) klein 1884]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2HH7 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2HH7 FirstGlance]. <br> | ||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=CU1:COPPER+(I)+ION'>CU1</scene></td></tr> | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CU1:COPPER+(I)+ION'>CU1</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2hh7 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2hh7 OCA], [https://pdbe.org/2hh7 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2hh7 RCSB], [https://www.ebi.ac.uk/pdbsum/2hh7 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2hh7 ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
[[ | [[https://www.uniprot.org/uniprot/CSOR_MYCTU CSOR_MYCTU]] Copper-sensitive repressor that has a key role in copper homeostasis. It is part of the cso operon involved in the cellular response to increasing concentrations of copper inside the bacterium, which can be highly toxic. In the presence of copper, CsoR fully dissociates from the promoter in the cso operon, leading to the transcription of its genes. Binds to a GC-rich pseudopallindromic sequence, 5'-GTAGCCCACCCCCAGTGGGGTGGGA-3', in the cso promoter region.<ref>PMID:17143269</ref> | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 31: | Line 31: | ||
==See Also== | ==See Also== | ||
*[[Copper homeostasis protein|Copper homeostasis protein]] | *[[Copper homeostasis protein|Copper homeostasis protein]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 14:08, 5 January 2022
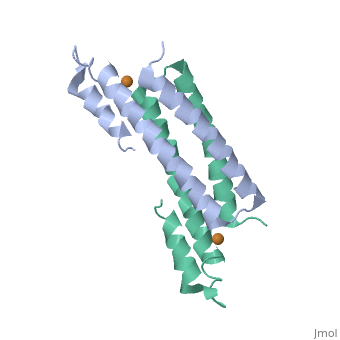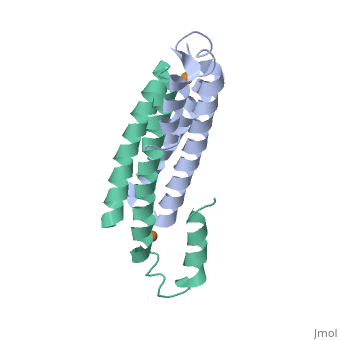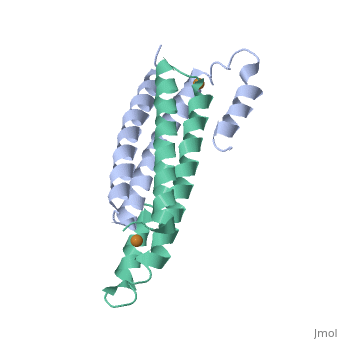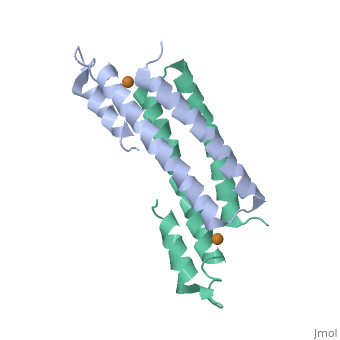
Crystal Structure of Cu(I) bound CsoR from Mycobacterium tuberculosis.Crystal Structure of Cu(I) bound CsoR from Mycobacterium tuberculosis.
Structural highlights
Function[CSOR_MYCTU] Copper-sensitive repressor that has a key role in copper homeostasis. It is part of the cso operon involved in the cellular response to increasing concentrations of copper inside the bacterium, which can be highly toxic. In the presence of copper, CsoR fully dissociates from the promoter in the cso operon, leading to the transcription of its genes. Binds to a GC-rich pseudopallindromic sequence, 5'-GTAGCCCACCCCCAGTGGGGTGGGA-3', in the cso promoter region.[1] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedCopper is an essential element that becomes highly cytotoxic when concentrations exceed the capacity of cells to sequester the ion. Here, we identify a new copper-specific repressor (CsoR) of a copper-sensitive operon (cso) in Mycobacterium tuberculosis (Mtb) that is representative of a large, previously uncharacterized family of proteins (DUF156). Electronic and X-ray absorption spectroscopies reveal that CsoR binds a single-monomer mole equivalent of Cu(I) to form a trigonally coordinated (S(2)N) Cu(I) complex. The 2.6-A crystal structure of copper-loaded CsoR shows a homodimeric antiparallel four-helix bundle architecture that represents a novel DNA-binding fold. The Cu(I) is coordinated by Cys36, Cys65' and His61' in a subunit bridging site. Cu(I) binding negatively regulates the binding of CsoR to a DNA fragment encompassing the operator-promoter region of the Mtb cso operon; this results in derepression of the operon in Mtb and the heterologous host Mycobacterium smegmatis. Substitution of Cys36 or His61 with alanine abolishes Cu(I)- and CsoR-dependent regulation in vivo and in vitro. Potential roles of CsoR in Mtb pathogenesis are discussed. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator.,Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP Nat Chem Biol. 2007 Jan;3(1):60-8. Epub 2006 Dec 3. PMID:17143269[2] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||