1rl6: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
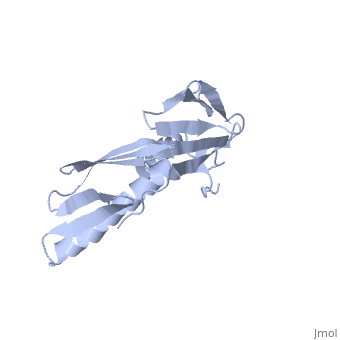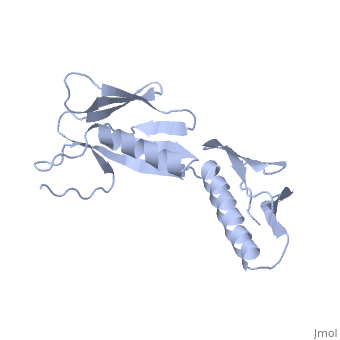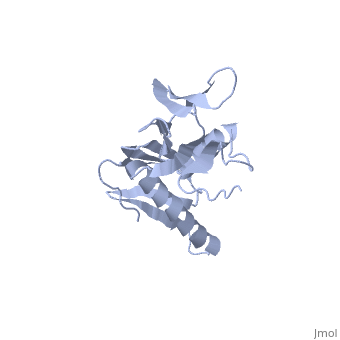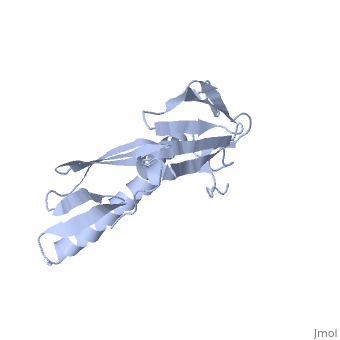
<StructureSection load='1rl6' size='340' side='right'caption='[[1rl6]], [[Resolution|resolution]] 2.00Å' scene=''> | <StructureSection load='1rl6' size='340' side='right'caption='[[1rl6]], [[Resolution|resolution]] 2.00Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1rl6]] is a 1 chain structure with sequence from [ | <table><tr><td colspan='2'>[[1rl6]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Atcc_12980 Atcc 12980]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1RL6 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1RL6 FirstGlance]. <br> | ||
</td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1rl6 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1rl6 OCA], [https://pdbe.org/1rl6 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1rl6 RCSB], [https://www.ebi.ac.uk/pdbsum/1rl6 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1rl6 ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
[[ | [[https://www.uniprot.org/uniprot/RL6_GEOSE RL6_GEOSE]] It is located near the subunit interface in the base of the L7/L12 stalk, and near the tRNA binding site of the peptidyltransferase center (By similarity). This protein binds to the 23S rRNA, and is important in its secondary structure.[HAMAP-Rule:MF_01365] | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Revision as of 13:14, 15 September 2021
RIBOSOMAL PROTEIN L6RIBOSOMAL PROTEIN L6
Structural highlights
Function[RL6_GEOSE] It is located near the subunit interface in the base of the L7/L12 stalk, and near the tRNA binding site of the peptidyltransferase center (By similarity). This protein binds to the 23S rRNA, and is important in its secondary structure.[HAMAP-Rule:MF_01365] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedIn all cells, protein synthesis is coordinated by the ribosome, a large ribonucleoprotein particle that is composed of > 50 distinct protein molecules and several large RNA molecules. Here we present the crystal structure of ribosomal protein L6 from the thermophilic bacterium Bacillus stearothermophilus solved at 2.6 A resolution. L6 contains two domains with almost identical folds, implying that it was created by an ancient gene duplication event. The surface of the molecule displays several likely sites of interaction with other components of the ribosome. The RNA binding sites appear to be localized in the C-terminal domain whereas the N-terminal domain contains the potential sites for protein-protein interactions. The domain structure is homologous with several other ribosomal proteins and to a large family of eukaryotic RNA binding proteins. Ribosomal protein L6: structural evidence of gene duplication from a primitive RNA binding protein.,Golden BL, Ramakrishnan V, White SW EMBO J. 1993 Dec 15;12(13):4901-8. PMID:8262035[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences |
| ||||||||||||||