1ecr: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
<StructureSection load='1ecr' size='340' side='right'caption='[[1ecr]], [[Resolution|resolution]] 2.70Å' scene=''> | <StructureSection load='1ecr' size='340' side='right'caption='[[1ecr]], [[Resolution|resolution]] 2.70Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1ecr]] is a 3 chain structure with sequence from [ | <table><tr><td colspan='2'>[[1ecr]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Escherichia_coli_(strain_b) Escherichia coli (strain b)]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1ECR OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1ECR FirstGlance]. <br> | ||
</td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1ecr FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ecr OCA], [https://pdbe.org/1ecr PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1ecr RCSB], [https://www.ebi.ac.uk/pdbsum/1ecr PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1ecr ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
[[ | [[https://www.uniprot.org/uniprot/TUS_ECOLI TUS_ECOLI]] Trans-acting protein required for termination of DNA replication. Binds to DNA replication terminator sequences (terA to terF) to prevent the passage of replication forks. The termination efficiency will be affected by the affinity of this protein for the terminator sequence. | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Revision as of 14:08, 28 July 2021
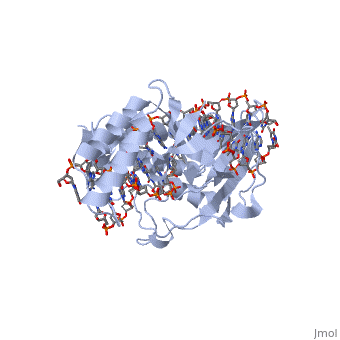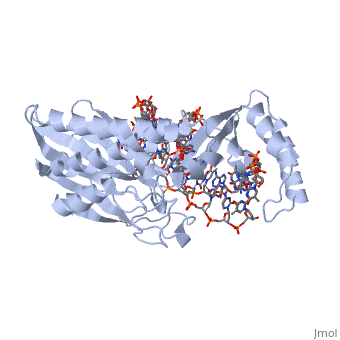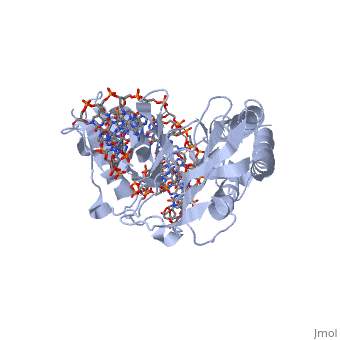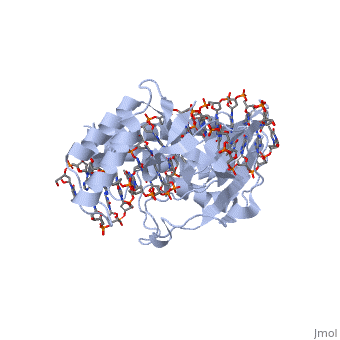
ESCHERICHIA COLI REPLICATION TERMINATOR PROTEIN (TUS) COMPLEXED WITH DNAESCHERICHIA COLI REPLICATION TERMINATOR PROTEIN (TUS) COMPLEXED WITH DNA
Structural highlights
Function[TUS_ECOLI] Trans-acting protein required for termination of DNA replication. Binds to DNA replication terminator sequences (terA to terF) to prevent the passage of replication forks. The termination efficiency will be affected by the affinity of this protein for the terminator sequence. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe crystal structure of the Escherichia coli replication-terminator protein (Tus) bound to terminus-site (Ter) DNA has been determined at 2.7 A resolution. The Tus protein folds into a previously undescribed architecture divided into two domains by a central basic cleft. This cleft accommodates locally deformed B-form Ter DNA and makes extensive contacts with the major groove, mainly through two interdomain beta-strands. The unusual structural features of this complex may explain how the replication fork is halted in only one direction. Structure of a replication-terminator protein complexed with DNA.,Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K Nature. 1996 Oct 17;383(6601):598-603. PMID:8857533[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||