Jmol/Cavities pockets and tunnels: Difference between revisions
Eric Martz (talk | contribs) No edit summary |
Eric Martz (talk | contribs) No edit summary |
||
| Line 94: | Line 94: | ||
==Ligands Inside Cavity== | ==Ligands Inside Cavity== | ||
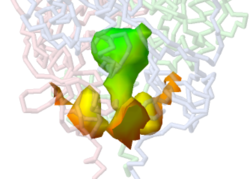
Bacterial glycosidase (alpha-N-acetylgalactosaminidase) 2ixb has a tunnel containing NAD that includes, at one entrance, a pocket where substrate can access the catalytic site. The tunnel contains both NAD and a substrate molecule. The following commands show the translucent tunnel with its contents rendered as sticks. | |||
:<tt>isosurface minset 100 select {protein} only pocket cavity property surfacedistance | Bacterial glycosidase (alpha-N-acetylgalactosaminidase) [[2ixb]] has a tunnel containing NAD that includes, at one entrance, a pocket where substrate can access the catalytic site. The tunnel contains both NAD and a substrate molecule. The following commands show the translucent tunnel with its contents rendered as sticks. | ||
:isosurface set 2</tt> | :<tt><scene name='85/858407/2ixb/1'>isosurface minset 100 select {protein} only pocket cavity property surfacedistance | ||
:isosurface set 2</scene></tt><!--2ixb-m100-poccav-sd-set1-xlu2.pngj--> | |||
*"select {protein} only" means that ligands and solvent are ignored during isosurface creation. | *"select {protein} only" means that ligands and solvent are ignored during isosurface creation. | ||
*"property surfacedistance" colors the surface by depth as part of the isosurface creation. | *"property surfacedistance" colors the surface by depth as part of the isosurface creation. | ||
Revision as of 00:49, 3 January 2021
CAUTION: This page is under construction and is not ready for use. When this page is adequately completed, this notice will be removed. Eric Martz 01:02, 1 January 2021 (UTC)
Jmol can find and display cavities, pockets, and tunnels as isosurfaces. This page explains how to do that, and how to show the results in Proteopedia. An alternative method for finding and displaying cavities is PACUPP. Several other methods are summarized at Cavity programs. TerminologyFor definitions of the terms cavity, pocket, tunnel, channel and void please see Cavity programs. How To Show Cavities In ProteopediaBelow are shown Jmol commands to identify and display cavities as isosurfaces, with their results. Using these commands, and displaying the results in Proeopedia, require several extra steps outside of Proteopedia's usual Scene Authoring Tools (SAT). In brief, cavity isosurfaces should be generated in the stand-alone Jmol application and captured "cached" in a PNGJ file, which is then uploaded to Proteopedia for use in the SAT. Instructions are at the end of this page under Preparing Isosurface Scenes for Proteopedia. You may also wish to consider an alternative method which represents cavities with pseudoatoms, PACUPP. Two ProbesJmol uses two "rolling" spherical probes to identify cavities.
Small Cavity ExampleThe coronavirus (SARS-CoV-2 and others) spike protein has a small, membrane proximal cavity (). It is a potential drug target. The simplest Jmol cavity command is This produces pockets, tunnels, and cavities of all sizes. There are 426 separate surfaces. The largest is a convoluted tunnel with many mouths, volume 60,197 Å3. Many are very tiny. Hiding Tiny Cavities: minsetSince we are not interested in the tiny ones, we can hide them by specifying a value for minset. Cavities rendered with fewer than the number of triangles specified for minset will not be shown. By trial and error, a good minset number is 1000, making the Jmol command: Now only the largest two cavities are shown. The larger one is a convoluted tunnel with many mouths, volume 60,197 Å3. The smaller one is the membrane-proximal cavity of interest, volume 7,591 Å3. Isolating One CavityThe command to display only the smaller cavity is: Coloring By DepthIt is revealing to color this cavity by distance from the surface with this command: [5] shows that the red parts, which are at or near the protein surface, are mostly closed. The "cavity" command, without modifiers, generates closed isosurfaces. Pockets vs. Interior CavitiesTo show the entrances or mouths of a cavity as open, the modifier pockets is needed. This modifier also hides cavities that lack openings to the macromolecular surface at the cavity probe radius employed.
Here is the command to show our cavity-of-interest's entrances from the protein surface as open: Spike protein is a homotrimer. The membrane-proximal cavity has three narrow tunnels, between protein chains, to wider pockets opening to the protein surface. Cavity Volumes: Closed OnlyJmol is not able to calculate a meaningful volume for a pocket rendered with open entrances. Volumes reported by Jmol for its closed rendering of the membrane-proximal cavity of the spike protein 6zgi are in rough agreement with those independently determined by PACUPP -- best at cavity probe radii of 1.5 or 2.0 Å.
Increasing the Cavity Probe RadiusAs the cavity probe radius increases, cavities are displayed with less detail. Also, cavities or tunnel connections between cavities that are smaller than the probe may fail to be detected. Recall that the deeper chamber of our membrane-proximal cavity of interest is connected to three entrance pockets by narrow tunnels. As the cavity probe radius increases, those narrow tunnels are no longer detected. At 2.0 Å, two of the entrances are no longer connected to the deeper chamber. At 2.5 Å, none are connected. The cavity probe radius in Å can be added to the end of Jmol's isosurface command like this: Because the deeper chamber has no entrances from the surface, it will not be shown when the entrance pockets are rendered open with the pockets modifier. Those open pockets must be combined with a separate isosurface calculation for the interior chamber. The scripts for doing this are below. Ligands Inside CavityBacterial glycosidase (alpha-N-acetylgalactosaminidase) 2ixb has a tunnel containing NAD that includes, at one entrance, a pocket where substrate can access the catalytic site. The tunnel contains both NAD and a substrate molecule. The following commands show the translucent tunnel with its contents rendered as sticks.
|
| |||||||||||||||||||||||||
Preparing Isosurface Scenes for ProteopediaPreparing Isosurface Scenes for Proteopedia
This section is under construction and awaits major revisions. It is not ready for use.
Speed of RenderingSpeed of Rendering
If you use the isosurface commands below, do so in the Jmol Java application, not in JSmol in Proteopedia. Depending on the size of the molecule, cavity isosurface commands take about a minute to complete in the Java application, which is many times faster than JSmol. You would have to wait many minutes for completion in JSmol.
Working DirectoryWorking Directory
Put a copy of Jmol.jar, and perhaps a downloaded copy of the PDB file(s) of interest in a new working directory (folder). By default, JVXL and PNGJ files you write will be written into the directory from which you start Jmol.jar. Any Jmol command script files should also be placed in this working directory. This keeps everything simple.
Use PNGJ FilesUse PNGJ Files
The best way to include cavity isosurfaces in a Proteopedia scene (green link) is to upload a properly prepared PNGJ file that includes the cached isosurfaces. Proteopedia's Molecular Scene Authoring Tools (SAT) load molecule tab accepts an uploaded PNGJ file.
Before saving the scene, you can add a caption that will display under the molecular display. The caption can include
- Links to pages within Proteopedia using double brackets, such as 6zgi (wikitext [[6zgi]]).
- Links to pages outside Proteopedia using single brackets, such as jmol.org (wikitext [http://jmol.org jmol.org]).
- Color keys using colored text. Coloring of text in captions must be done using span classes: see instructions at Coloring text. (In this example: <span class="text-red"><b>colored</b></span> <span class="bg-yellow">text</span>.)
Multiple IsosurfacesMultiple Isosurfaces
There are two reasons why you may need multiple isosurfaces.
- You want to show both closed interior cavities and open-mouthed surface pockets.
- You want to show multiple pieces of a single isosurface.
At present, Jmol can show only one piece of an isosurface. For example, above are shown three pocket entrances to the interior chamber of the spike protein cavity. Each pocket is displayed with an isosurface set <integer> command. But only one can be shown at a time. To display all 3, the JVXL file (see below) must be loaded 3 times, giving each a different ID, and then one pocket displayed from each. To also display the interior closed chamber, a 4th isosurface must be loaded. Below are scripts illustrating this.
Preparing PNGJ Files With Cached IsosurfacesPreparing PNGJ Files With Cached Isosurfaces
It is important to cache the isosurface(s) in your scene before saving the PNGJ file so the scene can load rapidly. If the PNGJ file is saved without caching, JSmol will re-calculate the isosurface(s) before the scene is displayed. This could easily take 10 minutes!
Here are the steps:
I. In the Jmol Java application:
- 1. Calculate the isosurface(s). Commands are explained above in earlier sections of this article. If you will need more than one isosurface, for example one for pockets and one for interior cavities, assign each a distinct ID when you create them. Prefixing the ID with tilde (~) ensures the ID name does not conflict with an existing term in Jmol. In a Jmol command line, anything after # is a comment, ignored by Jmol. For example:
- isosurface ~poc minset 100 pocket cavity
- isosurface ~bur minset 100 interior cavity # "buried"
- 2. Color the isosurface(s). Coloring by depth from the surface was described above. For a plain color, first you must know the ID of the isosurface if there are more than one.
- isosurface ~poc # Select the isosurface to color. Unnecessary if there is only one.
- color isosurface pink # color name
- isosurface ~bur # Select a different isosurface to color.
- color isosurface [x4060FF] # RGB color
- 3. Write each isosurface as a jvxl (Jmol Voxel) file. Coloring is saved in the JVXL file.
- isosurface ~poc # Select the isosurface to write. Unnecessary if there is only one.
- write poc.jvxl
- isosurface ~bur # Select a different isosurface to color.
- write bur.jvxl
- 4. Quit Jmol and start a new session. Load the molecule and then the JVXL file(s). If you are loading more than one JVXL file (or the same JVXL file more than once so you can display multiple pieces of it), assign each load a distinct ID. (ID's are not saved in the jvxl files.)
- load =6zgi
- isosurface ~poc1 poc.jvxl
- isosurface ~poc2 poc.jvxl
- isosurface ~poc3 poc.jvxl
- isosurface ~bur bur.jvxl
- 5. Cache the isosurface(s). This is critical. If this step is omitted, the PNGJ file will try to read the isosurfaces from the JVXL files, which will fail in Proteopedia.
- isosurface cache # A single command caches ALL isosurfaces.
- 6. Write the PNGJ file. It is best if you have rendered and colored the macromolecule exactly as you want the scene to appear in Proteopedia before writing the PNGJ file.
- write filename.pngj
II. In Proteopedia:
- 7. Upload your PNGJ file. You do NOT need to compress the PNGJ file because it is already in a compressed format. See Help:Uploading molecules. Keep a copy of the name of the page for your uploaded PNGJ file. For example, Image:6zgi-m1000-cav.pngj.
- 8. Now you are ready to use the SAT in your Proteopedia page. See Proteopedia:How to Make a Page. Use the load molecule tab in the SAT and paste in the name of the page containing your uploaded PNGJ file, such as Image:6zgi-m1000-cav.pngj. Use the save scene tab. Add a caption, and save the scene, creating a green link.
Working Example ScriptsWorking Example Scripts
The following script generates a pocket isosurface (open mouths) and an interior isosurface (closed pieces) for the spike protein, and saves each isosurface as a jvxl file. You can run this script: copy it and save it as a plain text file with a filename ending .spt. Drop the script file into the molecular display window of Jmol. Execution may take several minutes.
Write JVXL FilesWrite JVXL Files
# In the Jmol Java application, # This script took 2 min 25 sec on a mid-2014 MacBook Pro. # It would likely take ~20 min in JSmol. zap # Deletes any molecule and all isosurfaces present in Jmol. print # Clears the Jmol Script Console. load =6zgi # Fetches PDB file from the RCSB Protein Data Bank. # RENDER MOLECULE. Unnecessary but takes no time and looks nice while executing. background white rotate x -90 restrict protein # Selects protein and hides non-protein, e.g. glycosylation. backbone only backbone 0.7 color chain set translucent false # Shows only the front-most surface of translucent objects. color translucent 5 # Chain-colored backbones are now translucent. refresh # Unnecessary, but shows the molecule. # MAKE POCKETS isosurface minset 50 pocket cavity 2.5 # Cavity probe radius 2.5 Å. refresh isosurface map property surfacedistance # Color pockets by depth. refresh write "poc.jvxl" isosurface delete # MAKE INTERIOR CAVITY NOT DISPLAYED IN THE POCKETS ISOSURFACE. isosurface minset 100 interior cavity 2.5 refresh isosurface map property surfacedistance refresh write "bur.jvxl" # Buried.
Write Cached PNGJ FileWrite Cached PNGJ File
# Execution of this script took 4 seconds on a mid-2014 MacBook Pro.
zap # Deletes any molecule and all isosurfaces present in Jmol.
print # Clears the Jmol Script Console.
load =6zgi # Fetches PDB file from the RCSB Protein Data Bank.
# RENDER MOLECULE
background white
rotate x -90
restrict protein # Selects protein and hides non-protein, e.g. glycosylation.
backbone only
backbone 0.7
color chain
set translucent false # Shows only the front-most surface of translucent objects.
color translucent 5 # Chain-colored backbones are now translucent.
refresh # Unnecessary, but shows the molecule.
# DISPLAY OPEN POCKETS. Can display only one piece (set) per isosurface.
# To display three pieces (sets), create 3 (identical) isosurfaces.
# Set numbers determined by trial and error.
isosurface poc1 poc.jvxl
isosurface poc1 set 8
isosurface poc2 poc.jvxl
isosurface poc2 set 9
isosurface poc3 poc.jvxl
isosurface poc3 set 11
# CLOSED DEEPER CHAMBER.
isosurface bur bur.jvxl
isosurface bur set 1 # Unnecessary because set 1 is the only one passing minset 100.
delay 1.0 # shows the molecule with isosurfaces for 1 second.
# Arg1107 surrounds the deeper chamber.
# This centers that region, and zooms to 266%.
zoomto {arg1107} 266
isosurface cache # Without this, JSmol will fail trying to read the jvxl files absent in Proteopedia.
write "6zgi-pocbur2.5.pngj" # Ready to upload to Proteopedia.
Managing Cavity IsosurfacesManaging Cavity Isosurfaces
Jmol commands for generating cavity isosurfaces were explained above. Full details will be found in the Jmol/JSmol Interacive Scripting Documention under isosurfaces: molecular/solvent surfaces. Near the bottom of that very long section, important commands for after the cavity isosurfaces are calculated:
- isosurface area set (integer) # Reports the surface area of one piece (set) of an isosurface in Å2.
- isosurface * delete # Deletes all isosurfaces.
- isosurface set (integer) # Displays just one piece (set) of an isosurface.
- isosurface set 0 # Displays all pieces of an isosurface = the entire isosurface.
- isosurface volume set (integer) # Reports the volume of one closed[12] isosurface in Å3.
See AlsoSee Also
- Cavity programs
- PACUPP: Pockets And Cavities Using Pseudoatoms in Proteins
References and NotesReferences and Notes
- ↑ Bondi, A., J. Phys. Chem. 68:441, 1964.
- ↑ Diameter of water molecule at B10NUMB3R5, a collaboration between Harvard Medical School and the Weizmann Institute of Science. See also Distance between water molecules in bulk water.
- ↑ Volume of water molecule bound to an amino acid at B10NUMB3R5, a collaboration between Harvard Medical School and the Weizmann Institute of Science.
- ↑ This command works only if a previous isosurface ... cavity command has been run in the same session. It will not work if the surface is loaded from a JVXL file alone, because then the surface has not been defined.
- ↑ The command for close examination is based on finding that Asn1107 is close to the deep chamber of the membrane-proximal cavity. The command is zoomto {1107} 266. This centers the deep part of the cavity and zooms to 266%.
- ↑ 6.0 6.1 Cavity of interest was manually disconnected from the largest cavity.
- ↑ Sum of volumes of cavity sets 5 and 11.
- ↑ Sum of volumes of 2 clusters of pseudoatoms.
- ↑ Sum of volumes of cavity sets 6, 9, 10, and 12.
- ↑ Sum of volumes of 2 clusters of pseudoatoms. One quarter of the cavity failed to be detected with a cavity probe radius of 2.5 Å.
- ↑ PACUPP in offset mode fails to find 3/4 of the cavity with a cavity probe radius of 2.5 Å.
- ↑ The volume that Jmol reports for a pocket (open mouth/entrance) is meaningless. Jmol reports meaningful volumes only for closed isosurfaces.