2g2u: Difference between revisions
No edit summary |
No edit summary |
||
| Line 5: | Line 5: | ||
|SITE= | |SITE= | ||
|LIGAND= | |LIGAND= | ||
|ACTIVITY= [http://en.wikipedia.org/wiki/Beta-lactamase Beta-lactamase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.5.2.6 3.5.2.6] | |ACTIVITY= <span class='plainlinks'>[http://en.wikipedia.org/wiki/Beta-lactamase Beta-lactamase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.5.2.6 3.5.2.6] </span> | ||
|GENE= bla, shv1 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=573 Klebsiella pneumoniae]) | |GENE= bla, shv1 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=573 Klebsiella pneumoniae]) | ||
|DOMAIN= | |||
|RELATEDENTRY=[[2g2w|2G2W]] | |||
|RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2g2u FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2g2u OCA], [http://www.ebi.ac.uk/pdbsum/2g2u PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=2g2u RCSB]</span> | |||
}} | }} | ||
| Line 38: | Line 41: | ||
[[Category: shv-1]] | [[Category: shv-1]] | ||
''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 03:11:02 2008'' | ||
Revision as of 03:11, 31 March 2008
| |||||||
| , resolution 1.60Å | |||||||
|---|---|---|---|---|---|---|---|
| Gene: | bla, shv1 (Klebsiella pneumoniae) | ||||||
| Activity: | Beta-lactamase, with EC number 3.5.2.6 | ||||||
| Related: | 2G2W
| ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
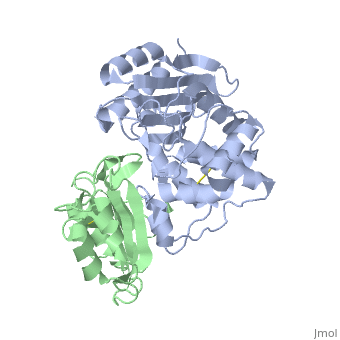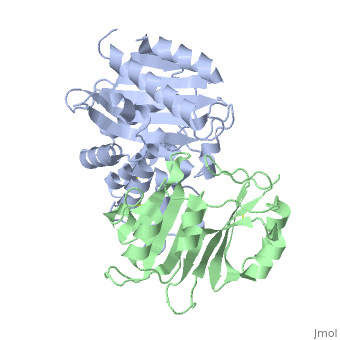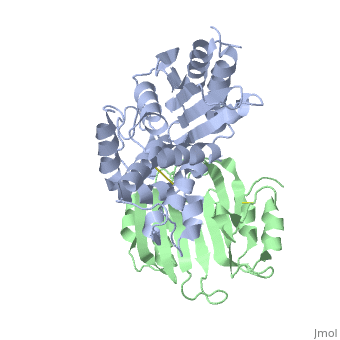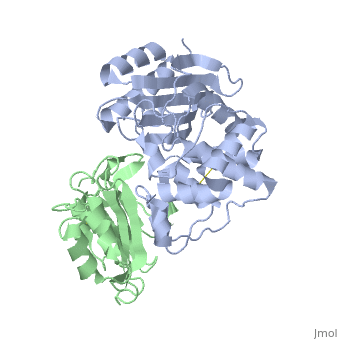
Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex
OverviewOverview
Beta-lactamase inhibitor protein (BLIP) binds a variety of class A beta-lactamases with affinities ranging from micromolar to picomolar. Whereas the TEM-1 and SHV-1 beta-lactamases are almost structurally identical, BLIP binds TEM-1 approximately 1000-fold tighter than SHV-1. Determining the underlying source of this affinity difference is important for understanding the molecular basis of beta-lactamase inhibition and mechanisms of protein-protein interface specificity and affinity. Here we present the 1.6A resolution crystal structure of SHV-1.BLIP. In addition, a point mutation was identified, SHV D104E, that increases SHV.BLIP binding affinity from micromolar to nanomolar. Comparison of the SHV-1.BLIP structure with the published TEM-1.BLIP structure suggests that the increased volume of Glu-104 stabilizes a key binding loop in the interface. Solution of the 1.8A SHV D104K.BLIP crystal structure identifies a novel conformation in which this binding loop is removed from the interface. Using these structural data, we evaluated the ability of EGAD, a program developed for computational protein design, to calculate changes in the stability of mutant beta-lactamase.BLIP complexes. Changes in binding affinity were calculated within an error of 1.6 kcal/mol of the experimental values for 112 mutations at the TEM-1.BLIP interface and within an error of 2.2 kcal/mol for 24 mutations at the SHV-1.BLIP interface. The reasonable success of EGAD in predicting changes in interface stability is a promising step toward understanding the stability of the beta-lactamase.BLIP complexes and computationally assisted design of tight binding BLIP variants.
About this StructureAbout this Structure
2G2U is a Protein complex structure of sequences from Klebsiella pneumoniae and Streptomyces clavuligerus. Full crystallographic information is available from OCA.
ReferenceReference
Structural and computational characterization of the SHV-1 beta-lactamase-beta-lactamase inhibitor protein interface., Reynolds KA, Thomson JM, Corbett KD, Bethel CR, Berger JM, Kirsch JF, Bonomo RA, Handel TM, J Biol Chem. 2006 Sep 8;281(36):26745-53. Epub 2006 Jun 29. PMID:16809340
Page seeded by OCA on Mon Mar 31 03:11:02 2008