1emv: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
==CRYSTAL STRUCTURE OF COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 (1.7 ANGSTROMS)== | ==CRYSTAL STRUCTURE OF COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 (1.7 ANGSTROMS)== | ||
<StructureSection load='1emv' size='340' side='right' caption='[[1emv]], [[Resolution|resolution]] 1.70Å' scene=''> | <StructureSection load='1emv' size='340' side='right' caption='[[1emv]], [[Resolution|resolution]] 1.70Å' scene=''> | ||
| Line 6: | Line 7: | ||
<tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1bxi|1bxi]]</td></tr> | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1bxi|1bxi]]</td></tr> | ||
<tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Deoxyribonuclease_I Deoxyribonuclease I], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.1.21.1 3.1.21.1] </span></td></tr> | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Deoxyribonuclease_I Deoxyribonuclease I], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.1.21.1 3.1.21.1] </span></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1emv FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1emv OCA], [http://pdbe.org/1emv PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=1emv RCSB], [http://www.ebi.ac.uk/pdbsum/1emv PDBsum]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1emv FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1emv OCA], [http://pdbe.org/1emv PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=1emv RCSB], [http://www.ebi.ac.uk/pdbsum/1emv PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=1emv ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| Line 14: | Line 15: | ||
Check<jmol> | Check<jmol> | ||
<jmolCheckbox> | <jmolCheckbox> | ||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/em/1emv_consurf.spt"</scriptWhenChecked> | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/em/1emv_consurf.spt"</scriptWhenChecked> | ||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
| Line 29: | Line 30: | ||
</div> | </div> | ||
<div class="pdbe-citations 1emv" style="background-color:#fffaf0;"></div> | <div class="pdbe-citations 1emv" style="background-color:#fffaf0;"></div> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 10:03, 20 December 2017
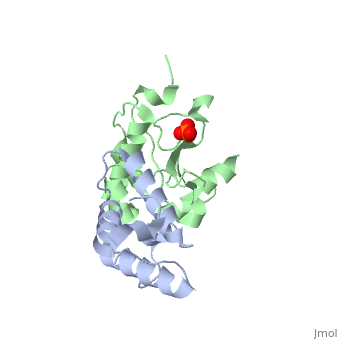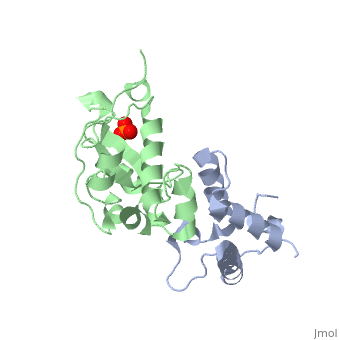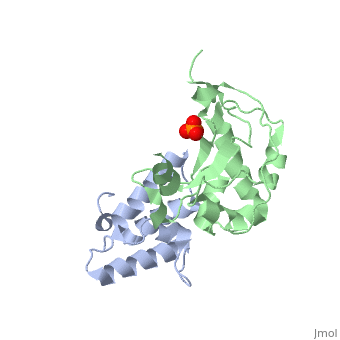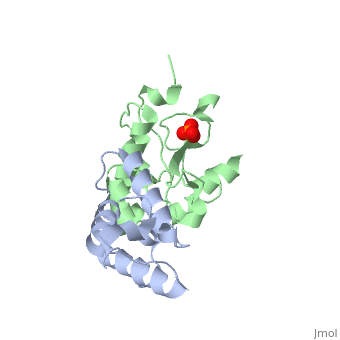
CRYSTAL STRUCTURE OF COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 (1.7 ANGSTROMS)CRYSTAL STRUCTURE OF COLICIN E9 DNASE DOMAIN WITH ITS COGNATE IMMUNITY PROTEIN IM9 (1.7 ANGSTROMS)
Structural highlights
Function[IMM9_ECOLX] This protein is able to protect a cell, which harbors the plasmid ColE9 encoding colicin E9, against colicin E9, it binds specifically to the DNase-type colicin and inhibits its bactericidal activity. [CEA9_ECOLX] This plasmid-coded bactericidal protein is an endonuclease active on both single- and double-stranded DNA but with undefined specificity. Colicins are polypeptide toxins produced by and active against E.coli and closely related bacteria. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedBacteria producing endonuclease colicins are protected against their cytotoxic activity by virtue of a small immunity protein that binds with high affinity and specificity to inactivate the endonuclease. DNase binding by the immunity protein occurs through a "dual recognition" mechanism in which conserved residues from helix III act as the binding-site anchor, while variable residues from helix II define specificity. We now report the 1.7 A crystal structure of the 24.5 kDa complex formed between the endonuclease domain of colicin E9 and its cognate immunity protein Im9, which provides a molecular rationale for this mechanism. Conserved residues of Im9 form a binding-energy hotspot through a combination of backbone hydrogen bonds to the endonuclease, many via buried solvent molecules, and hydrophobic interactions at the core of the interface, while the specificity-determining residues interact with corresponding specificity side-chains on the enzyme. Comparison between the present structure and that reported recently for the colicin E7 endonuclease domain in complex with Im7 highlights how specificity is achieved by very different interactions in the two complexes, predominantly hydrophobic in nature in the E9-Im9 complex but charged in the E7-Im7 complex. A key feature of both complexes is the contact between a conserved tyrosine residue from the immunity proteins (Im9 Tyr54) with a specificity residue on the endonuclease directing it toward the specificity sites of the immunity protein. Remarkably, this tyrosine residue and its neighbour (Im9 Tyr55) are the pivots of a 19 degrees rigid-body rotation that relates the positions of Im7 and Im9 in the two complexes. This rotation does not affect conserved immunity protein interactions with the endonuclease but results in different regions of the specificity helix being presented to the enzyme. Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes.,Kuhlmann UC, Pommer AJ, Moore GR, James R, Kleanthous C J Mol Biol. 2000 Sep 1;301(5):1163-78. PMID:10966813[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. References
|
| ||||||||||||||||||||