Unusual sequence numbering: Difference between revisions
Eric Martz (talk | contribs) |
Eric Martz (talk | contribs) |
||
| Line 18: | Line 18: | ||
'''Negative.''' Sometimes the initial sequence number is negative. This is usually done when residues were engineered onto the N-terminus. The transition from -1 to 1 may or may not include a residue numbered zero. An example is [http://firstglance.jmol.org/fg.htm?mol=1d5t 1d5t] ([[1d5t]]). The N-terminal Met of the [http://www.uniprot.org/uniprot/P21856#sequences genomic sequence] is numbered 1. But a di-histidine tag was engineered onto the N-terminus: His -2, His -1, Met 1. In this case, there is no residue numbered zero. The C-terminal residue is Phe431, but the length of the genomic sequence is 447. The C-terminal 16 residues of the genomic sequence were not present in the crystallized protein. In this model, no residues are missing due to crystallographic disorder. | '''Negative.''' Sometimes the initial sequence number is negative. This is usually done when residues were engineered onto the N-terminus. The transition from -1 to 1 may or may not include a residue numbered zero. An example is [http://firstglance.jmol.org/fg.htm?mol=1d5t 1d5t] ([[1d5t]]). The N-terminal Met of the [http://www.uniprot.org/uniprot/P21856#sequences genomic sequence] is numbered 1. But a di-histidine tag was engineered onto the N-terminus: His -2, His -1, Met 1. In this case, there is no residue numbered zero. The C-terminal residue is Phe431, but the length of the genomic sequence is 447. The C-terminal 16 residues of the genomic sequence were not present in the crystallized protein. In this model, no residues are missing due to crystallographic disorder. | ||
Another example is [http://firstglance.jmol.org/fg.htm?mol=4ifd 4ifd] ([[4ifd]]), where chain R includes RNA residues numbered -1 to -10 and -30 to -44. | |||
==Multiple Residues with the Same Number== | ==Multiple Residues with the Same Number== | ||
Revision as of 22:24, 5 December 2017
The numbering of protein and nucleic acid sequences is arbitrary in structure files from the World Wide Protein Data Bank (PDB). That is, authors are free to number sequences as they wish.
Straightforward numbering assigns 1 to the amino-terminal amino acid, and counts up sequentially and monotonically to the carboxy-terminal amino acid. An example is 1pgb (1pgb). The crystallized protein is numbered 1-56, despite it being a fragment of a 448-residue full length sequence that begins (after adding an N-terminal Met) at full-length sequence number 228.
Below are some examples of unusual sequence numbering. The 3D structures of these PDB entries are not shown here. To explore them in 3D, the links below will display them in FirstGlance in Jmol (link with arrow) or in Proteopedia (link in parentheses).
Numbering Does Not Start With OneNumbering Does Not Start With One
N-Terminal Residues Missing CoordinatesN-Terminal Residues Missing Coordinates
Probably the most common reason that the first residue with coordinates is not numbered 1 is because the N-terminal (or 5'-terminal) residues are missing coordinates due to crystallographic disorder (fuzzy electron density map). An example is 1d66 (1d66). The first 7 residues of chain A are missing, so the first residue with coordinates is numbered 8. 1-7 were present in the crystallized protein, but could not be resolved in the electron density map.
N-Terminal Residues Deleted From ProteinN-Terminal Residues Deleted From Protein
Another common reason that sequence numbering does not start with 1 is because a range of N-terminal residues were deleted from the cloned and expressed protein used in the experiment. An example is chain A in 1b07 (1b07). This 65 amino acid chain starts with Gly132-Ser133 that are not part of the gene sequence. Next comes Ala134, and its sequence number (and the numbering of the remainder of the chain) matches the numbering of the gene-encoded protein, full length 304 amino acids.
Authors do not always use the full-length sequence numbering when the structure of a fragment is reported. As mentioned above, in 1pgb (1pgb), the crystallized protein is numbered 1-56. This despite it being a fragment of a 448-residue full length sequence that begins (after adding an N-terminal Met) at full-length sequence number 228.
Starts With Zero Or Negative NumbersStarts With Zero Or Negative Numbers
Zero. Sometimes the initial sequence number is zero. An example is 1bxw (1bxw). The first 21 residues of the genomic sequence are a signal sequence. The crystallized protein was engineered to start at residue 22 of the genomic sequence, which is Ala1 of the mature protein. A Met was engineered onto the N-terminus presumably to assist with expression. It was numbered Met0. (The crystallized protein ends at 178, but the length of the genomic sequence of the mature protein is 346 - 21 = 325.)
Negative. Sometimes the initial sequence number is negative. This is usually done when residues were engineered onto the N-terminus. The transition from -1 to 1 may or may not include a residue numbered zero. An example is 1d5t (1d5t). The N-terminal Met of the genomic sequence is numbered 1. But a di-histidine tag was engineered onto the N-terminus: His -2, His -1, Met 1. In this case, there is no residue numbered zero. The C-terminal residue is Phe431, but the length of the genomic sequence is 447. The C-terminal 16 residues of the genomic sequence were not present in the crystallized protein. In this model, no residues are missing due to crystallographic disorder.
Another example is 4ifd (4ifd), where chain R includes RNA residues numbered -1 to -10 and -30 to -44.
Multiple Residues with the Same NumberMultiple Residues with the Same Number
Insertion CodesInsertion Codes
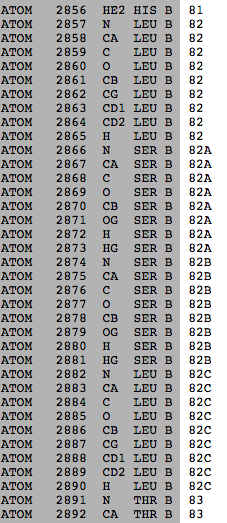
Sometimes the residues of a protein are numbered according to a different reference sequence. When there are insertions relative to the reference sequence, the additional residues may all be given the same sequence number, but marked with alphabetic insertion codes. This is frequently done in antibodies, where the reference sequence is the germline sequence, but the antibody has been somatically mutated, especially in complementarity-determining region (CDR) 3. An example is 1igy (1igy). Four residues in chain B all have sequence number 82. They are distinguished by insertion codes: 82, 82A, 82B, 82C. At right is this part of the PDB file.
Insertion Codes In ReverseInsertion Codes In Reverse
Rarely, the insertion codes are in reverse alphabetical order. An example is 1ucy (1ucy). Chain L begins with nine amino acids all numbered 1. The insertion codes are in reverse-alphabetic order: 1H, 1G, 1F, ... 1B, 1A, 1, 2, 3 .... In the same chain L are fourteen residues numbered 14. These insertion codes are in forward alphabetic order: 13, 14, 14A, 14B, ... 14L, 14M, 15, 16 .... Chain L also has ten residues numbered 60, with forward-alphabetic insertion codes from A through I, and a few other shorter runs of insertion codes.
Gaps In Sequence NumberingGaps In Sequence Numbering
Skipping Sequence NumbersSkipping Sequence Numbers
Sometimes a range of sequence numbers is skipped when numbering a continuous protein chain. There is no gap in the protein chain, but merely a discontinuity in the numbering of the chain. In the case of antibody 1igt (1igt), the sequence is numbered according to the Kabat scheme, relative to a reference sequence. Chain B begins with 1 and ends with 474 but contains only 444 residues (none are missing coordinates due to disorder). In chain B, residue 97 is followed by residue 100, skipping numbers 98-99. Only the numbers are skipped. No residues are missing. Residue 97 is peptide-bonded to residue 100. There are four residues 100, with insertion codes H, I, J, K. Residue 157 is followed by residue 162, skipping numbers 158-161. Also skipped are sequence numbers 170, 181-182, 197, 201, 207, 224-225, 233-234, 293-294, 297-298, 315-316, 356, 362, 376, 380, 403-404, 409, 412-413, 429, 431-432, and probably more.
Missing ResiduesMissing Residues
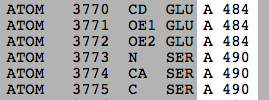
It is not uncommon for a surface loop of the crystallized protein to be disordered. Often such loops are intrinsically disordered. The disorder blurs the electron density map for that loop, and the loop residues are not given coordinates in the model: they are missing in the model. However, they were not missing in the crystallized protein. This causes a gap in the sequence numbers in the PDB file. An example is 2ace (2ace). Residues 485-489 are missing in the 3D crystallographic model due to disorder in the crystal. Also missing are 3 N-terminal, and 2 C-terminal residues. FirstGlance in Jmol tabulates missing residues, and marks regions of the 3D model where residues are missing with "empty baskets".
Not MonotonicNot Monotonic
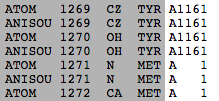
Rarely, sequence numbers do not increase monotonically from N to C terminus. An example[1] is 4zwj (4zwj). In this chimeric protein, chain A is numbered 1002-1161 continuing 1-326 continuing 2012-2361. That is, there are sudden jumps in numbering of consecutive amino acids: 1161 to 1, and 326 to 2012. At right is an excerpt from the ATOM records of the PDB file for 4zwj chain A.