3rif: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
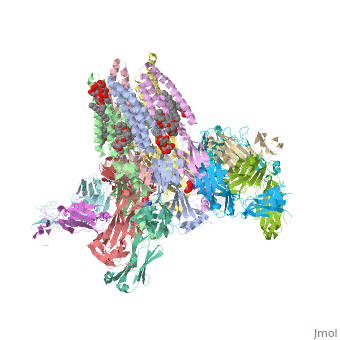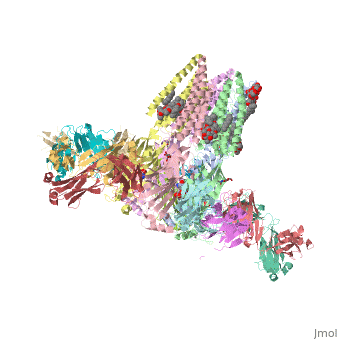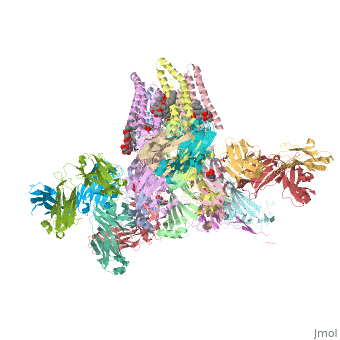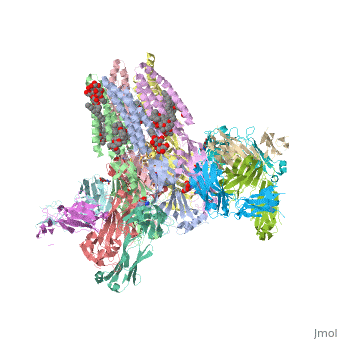
==C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and glutamate.== | ==C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and glutamate.== | ||
<StructureSection load='3rif' size='340' side='right' caption='[[3rif]], [[Resolution|resolution]] 3.34Å' scene=''> | <StructureSection load='3rif' size='340' side='right' caption='[[3rif]], [[Resolution|resolution]] 3.34Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[3rif]] is a 15 chain structure with sequence from [http://en.wikipedia.org/wiki/ | <table><tr><td colspan='2'>[[3rif]] is a 15 chain structure with sequence from [http://en.wikipedia.org/wiki/Caeel Caeel] and [http://en.wikipedia.org/wiki/Lk3_transgenic_mice Lk3 transgenic mice]. The November 2015 RCSB PDB [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Glutamate-gated Chloride Receptors'' by David Goodsell is [http://dx.doi.org/10.2210/rcsb_pdb/mom_2015_11 10.2210/rcsb_pdb/mom_2015_11]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3RIF OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3RIF FirstGlance]. <br> | ||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=GLU:GLUTAMIC+ACID'>GLU</scene>, <scene name='pdbligand=IVM:(2AE,4E,5S,6S,6R,7S,8E,11R,13R,15S,17AR,20R,20AR,20BS)-6-[(2S)-BUTAN-2-YL]-20,20B-DIHYDROXY-5,6,8,19-TETRAMETHYL-17-OXO-3,4,5,6,6,10,11,14,15,17,17A,20,20A,20B-TETRADECAHYDRO-2H,7H-SPIRO[11,15-METHANOFURO[4,3,2-PQ][2,6]BENZODIOXACYCLOOCTADECINE-13,2-PYRAN]-7-YL+2,6-DIDEOXY-4-O-(2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSYL)-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE'>IVM</scene>, <scene name='pdbligand=LMT:DODECYL-BETA-D-MALTOSIDE'>LMT</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand=OCT:N-OCTANE'>OCT</scene>, <scene name='pdbligand=UND:UNDECANE'>UND</scene></td></tr> | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=GLU:GLUTAMIC+ACID'>GLU</scene>, <scene name='pdbligand=IVM:(2AE,4E,5S,6S,6R,7S,8E,11R,13R,15S,17AR,20R,20AR,20BS)-6-[(2S)-BUTAN-2-YL]-20,20B-DIHYDROXY-5,6,8,19-TETRAMETHYL-17-OXO-3,4,5,6,6,10,11,14,15,17,17A,20,20A,20B-TETRADECAHYDRO-2H,7H-SPIRO[11,15-METHANOFURO[4,3,2-PQ][2,6]BENZODIOXACYCLOOCTADECINE-13,2-PYRAN]-7-YL+2,6-DIDEOXY-4-O-(2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSYL)-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE'>IVM</scene>, <scene name='pdbligand=LMT:DODECYL-BETA-D-MALTOSIDE'>LMT</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand=OCT:N-OCTANE'>OCT</scene>, <scene name='pdbligand=UND:UNDECANE'>UND</scene></td></tr> | ||
<tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3rhw|3rhw]], [[3ri5|3ri5]], [[3ria|3ria]]</td></tr> | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3rhw|3rhw]], [[3ri5|3ri5]], [[3ria|3ria]]</td></tr> | ||
<tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">glc-1, F11A5.10 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=6239 | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">glc-1, F11A5.10 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=6239 CAEEL])</td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3rif FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3rif OCA], [http://www.rcsb.org/pdb/explore.do?structureId=3rif RCSB], [http://www.ebi.ac.uk/pdbsum/3rif PDBsum]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3rif FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3rif OCA], [http://pdbe.org/3rif PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=3rif RCSB], [http://www.ebi.ac.uk/pdbsum/3rif PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=3rif ProSAT]</span></td></tr> | ||
</table> | </table> | ||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
| Line 16: | Line 17: | ||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
</div> | </div> | ||
<div class="pdbe-citations 3rif" style="background-color:#fffaf0;"></div> | |||
==See Also== | ==See Also== | ||
*[[ | *[[3D structures of monoclonal antibody|3D structures of monoclonal antibody]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
[[Category: | [[Category: Caeel]] | ||
[[Category: | [[Category: Glutamate-gated Chloride Receptors]] | ||
[[Category: Lk3 transgenic mice]] | |||
[[Category: RCSB PDB Molecule of the Month]] | |||
[[Category: Gouaux, E]] | [[Category: Gouaux, E]] | ||
[[Category: Hibbs, R E]] | [[Category: Hibbs, R E]] | ||
Revision as of 14:18, 10 December 2016
C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and glutamate.C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab, ivermectin and glutamate.
Structural highlights
Publication Abstract from PubMedFast inhibitory neurotransmission is essential for nervous system function and is mediated by binding of inhibitory neurotransmitters to receptors of the Cys-loop family embedded in the membranes of neurons. Neurotransmitter binding triggers a conformational change in the receptor, opening an intrinsic chloride channel and thereby dampening neuronal excitability. Here we present the first three-dimensional structure, to our knowledge, of an inhibitory anion-selective Cys-loop receptor, the homopentameric Caenorhabditis elegans glutamate-gated chloride channel alpha (GluCl), at 3.3 A resolution. The X-ray structure of the GluCl-Fab complex was determined with the allosteric agonist ivermectin and in additional structures with the endogenous neurotransmitter l-glutamate and the open-channel blocker picrotoxin. Ivermectin, used to treat river blindness, binds in the transmembrane domain of the receptor and stabilizes an open-pore conformation. Glutamate binds in the classical agonist site at subunit interfaces, and picrotoxin directly occludes the pore near its cytosolic base. GluCl provides a framework for understanding mechanisms of fast inhibitory neurotransmission and allosteric modulation of Cys-loop receptors. Principles of activation and permeation in an anion-selective Cys-loop receptor.,Hibbs RE, Gouaux E Nature. 2011 May 15. PMID:21572436[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||||