Sandbox Reserved 439: Difference between revisions
No edit summary |
No edit summary |
||
| Line 8: | Line 8: | ||
[[Student Projects for UMass Chemistry 423 Spring 2016]] | [[Student Projects for UMass Chemistry 423 Spring 2016]] | ||
<StructureSection load='1wat' size='350' side='right' caption='Bacteria use this protein to "smell" their environment (PDB entry [[1wat]])' scene=''> | <StructureSection load='1wat' size='350' side='right' caption='Bacteria use this protein to "smell" their environment (PDB entry [[1wat]])' scene=''> | ||
<font color=' | <font color='red'>This is an example page. Edit appropriately for your project. Upon completion of each task, delete instructions in red. See [[Sandbox Reserved 439]] for original instructions</font> | ||
<font color=' | <font color='red'>1. Add a citation to the primary reference at the end of the title above: you should all read this paper! Go to the [http://www.rcsb.org/pdb/home/home.do pdb] and search for your pdb code. Go down to the abstract and click "Search on Pubmed". | ||
In pubmed, copy the PMID code you see right below the abstract, and paste it to replace xxx after PMID in your title (first edited line above).</font> | In pubmed, copy the PMID code you see right below the abstract, and paste it to replace xxx after PMID in your title (first edited line above).</font> | ||
<font color='red'>** I can't figure out why the title citation gets repeated (refs 1 and 2) when it is cited in the text... We'll just live with it.**</font> | <font color='red'>** I can't figure out why the title citation gets repeated (refs 1 and 2) when it is cited in the text... We'll just live with it.**</font> | ||
<font color=' | <font color='red'>2. Each section should start with an "initial view" green scene. These can be the same for all sections or can be set by the section author. Section text should provide an interesting description (suitable for non-experts), and illustrate your points about the complex with multiple green scenes. Section text should not exceed the length of the structure window (<300 words). Do not include any copyrighted figures! All sections must be written in your own words with citations to your sources, following the reference format in this example (go into edit this page, to see how title reference was done). Make it interesting and accessible to a non-scientist! Show clearly some chemical details to illustrate the chemistry of life processes!</font> | ||
==Introduction== | ==Introduction== | ||
<font color=' | <font color='red'>Introduce the protein function, how it is related to a disease or fundamental process, and what is important about the ligand in the complex. Make a few green scenes suitable for the Molecular Playground: attractive and informative illustrations of (1) the protein complex, and (2) what is especially interesting about it. Think of a clever caption for potential display at the Molecular Playground -- caption must be short, ideally < 10 words. Edit the StructureSectionLoad command (4th edited line) from the top to insert your caption.</font> | ||
<scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>(Initial view)</scene> | <scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>(Initial view)</scene> | ||
Revision as of 22:11, 2 March 2016
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
Asp Receptor Ligand-binding domain (1wat)[1]Asp Receptor Ligand-binding domain (1wat)[1]
by [list all teammate names here] Student Projects for UMass Chemistry 423 Spring 2016
This is an example page. Edit appropriately for your project. Upon completion of each task, delete instructions in red. See Sandbox Reserved 439 for original instructions 1. Add a citation to the primary reference at the end of the title above: you should all read this paper! Go to the pdb and search for your pdb code. Go down to the abstract and click "Search on Pubmed". In pubmed, copy the PMID code you see right below the abstract, and paste it to replace xxx after PMID in your title (first edited line above). ** I can't figure out why the title citation gets repeated (refs 1 and 2) when it is cited in the text... We'll just live with it.** 2. Each section should start with an "initial view" green scene. These can be the same for all sections or can be set by the section author. Section text should provide an interesting description (suitable for non-experts), and illustrate your points about the complex with multiple green scenes. Section text should not exceed the length of the structure window (<300 words). Do not include any copyrighted figures! All sections must be written in your own words with citations to your sources, following the reference format in this example (go into edit this page, to see how title reference was done). Make it interesting and accessible to a non-scientist! Show clearly some chemical details to illustrate the chemistry of life processes! IntroductionIntroduce the protein function, how it is related to a disease or fundamental process, and what is important about the ligand in the complex. Make a few green scenes suitable for the Molecular Playground: attractive and informative illustrations of (1) the protein complex, and (2) what is especially interesting about it. Think of a clever caption for potential display at the Molecular Playground -- caption must be short, ideally < 10 words. Edit the StructureSectionLoad command (4th edited line) from the top to insert your caption.
The ligand binding domain of the aspartate receptor is a dimer of two 4-helix bundles that is shown here with the bound.[2] Overall Structure
In this the N and C termini are at the bottom of the structure; this is where the connections to the transmembrane helices have been truncated. Binding Interactions
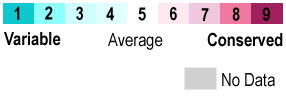
When the protein is colored according to , residues at the ligand site are the most conserved. Interactions that stabilize ligand binding[3] include hydrogen bonding from Tyr149 and Gln152 backbone carbonyls and Thr154 sidechain OH to the and hydrogen bonding from the sidechain nitrogens of Arg64, Arg69, and Arg73 to the two . Additional Features
Quiz Question 1
See AlsoCreditsIntroduction - name of team member Overall Structure - name of team member Binding Interactions - name of team member Additional Features - name of team member Quiz Question 1 - name of team member References
|
| ||||||||||