Sandbox Reserved 428: Difference between revisions
| Line 3: | Line 3: | ||
<!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | <!-- INSERT YOUR SCENES AND TEXT BELOW THIS LINE --> | ||
=='''Vitamin D receptor/vitamin D (1db1)'''== | =='''Vitamin D receptor/vitamin D (1db1)<ref>PMID: 10678179 </ref>'''== | ||
by Roger Crocker, Kate Daborowski, Patrick Murphy, Benjamin Rizkin and Aaron Thole | by Roger Crocker, Kate Daborowski, Patrick Murphy, Benjamin Rizkin and Aaron Thole | ||
Revision as of 21:02, 25 February 2016
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
Vitamin D receptor/vitamin D (1db1)[1]Vitamin D receptor/vitamin D (1db1)[1]
by Roger Crocker, Kate Daborowski, Patrick Murphy, Benjamin Rizkin and Aaron Thole
Student Projects for UMass Chemistry 423 Spring 2016
IntroductionIntroduction
|
What to talk about:
What class of proteins it belongs to, how its characterized, other names
-Nuclear hormone receptor
-Component of Mediator complex
Basic function
-transcription regulator of hormone sensitive genes
-has central role in calcium homeostasis
Retailed Diseases
-Rickets, hypocalcemia, secondary hyperparathyroidism, total alopecia
Importance
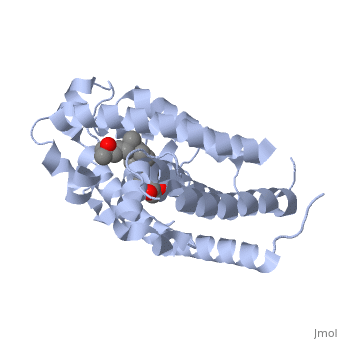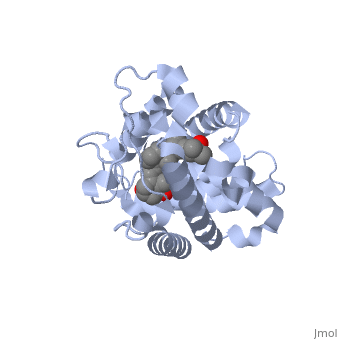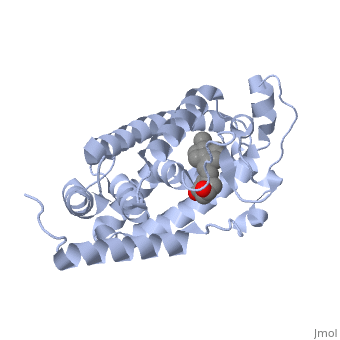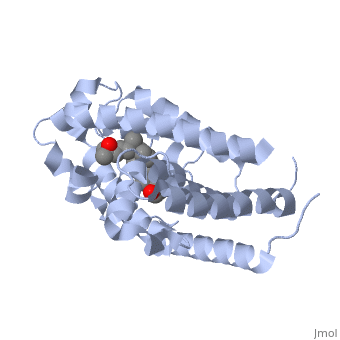
Overall StructureOverall Structure
|
Outline: -Discuss Primary Structure[ -Discuss Secondary Structure -Comprised of alpha helices and 1 beta sheet -Discuss tertiary structure -No quaternary structure -427 amino acids, 48289 Da (http://www.genecards.org/cgi-bin/carddisp.pl?gene=VDR)
Binding InteractionsBinding Interactions
|
Protein 1db1 is found to complex with 1,25 Dihydroxy . This molecule has three notable alcohol groups that have the ability to participate in interactions with the protein. Vitamin D3 has a large number of relations with the residues on the protein chain. First in the sequence are . Tyr143, shown in blue, is the closest to the ligand at 2.83 angstroms. This is sightly large but there is still the possibility of hydrogen bonding. Tyr147 in green and Phe150 in black are also known to have interactions with Vitamin D3 they are farther away and therefore less significant. Outline: as has been started, the interactions of the various residues will be covered. The types of bonds that 1db1 and VDX form will be elaborated as well
Additional FeaturesAdditional Features
|
A mutation in the transcription of the protein has the potential to result in the disease known as type 2 rickets. The mutation results in the not forming properly. Therefore VDR cannot complex with VDX. Outline: -Discuss mutations in Vitamin D receptor (Rickets) -Regulating hair cycle -Examine attempts to regulate Vitamin D receptor
Quiz Question 1Quiz Question 1
|
Quiz question: More information is needed to focus what the quiz should be about. Possibly related to how the secondary structure is comprised almost entirely of alpha helices, and how that relates to the amino acid sequences.
See AlsoSee Also
CreditsCredits
Introduction - Kate Daborowski
Overall Structure - Aaron Thole and Benjamin Rizkin
Drug Binding Site - Roger Crocker
Additional Features - Patrick Murphy
Quiz Question 1 - name of team member