2ysu: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
<StructureSection load='2ysu' size='340' side='right' caption='[[2ysu]], [[Resolution|resolution]] 3.50Å' scene=''> | <StructureSection load='2ysu' size='340' side='right' caption='[[2ysu]], [[Resolution|resolution]] 3.50Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[2ysu]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/ | <table><tr><td colspan='2'>[[2ysu]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/"bacillus_coli"_migula_1895 "bacillus coli" migula 1895]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2YSU OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2YSU FirstGlance]. <br> | ||
</td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2ysu FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2ysu OCA], [http://www.rcsb.org/pdb/explore.do?structureId=2ysu RCSB], [http://www.ebi.ac.uk/pdbsum/2ysu PDBsum]</span></td></tr> | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=2ysu FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2ysu OCA], [http://pdbe.org/2ysu PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=2ysu RCSB], [http://www.ebi.ac.uk/pdbsum/2ysu PDBsum]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
[[http://www.uniprot.org/uniprot/BTUB_ECOLI BTUB_ECOLI]] Involved in the active translocation of vitamin B12 (cyanocobalamin) across the outer membrane to the periplasmic space. It derives its energy for transport by interacting with the trans-periplasmic membrane protein TonB. Is also a receptor for bacteriophages BF23 and C1, and for A and E colicins.[HAMAP-Rule:MF_01531] | [[http://www.uniprot.org/uniprot/BTUB_ECOLI BTUB_ECOLI]] Involved in the active translocation of vitamin B12 (cyanocobalamin) across the outer membrane to the periplasmic space. It derives its energy for transport by interacting with the trans-periplasmic membrane protein TonB. Is also a receptor for bacteriophages BF23 and C1, and for A and E colicins.[HAMAP-Rule:MF_01531] [[http://www.uniprot.org/uniprot/CEA2_ECOLX CEA2_ECOLX]] This plasmid-coded bactericidal protein is an endonuclease active on both single- and double-stranded DNA but with undefined specificity. Colicins are polypeptide toxins produced by and active against E.coli and closely related bacteria. | ||
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 15: | Line 15: | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/ | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2ysu ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
| Line 25: | Line 25: | ||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
</div> | </div> | ||
<div class="pdbe-citations 2ysu" style="background-color:#fffaf0;"></div> | |||
==See Also== | ==See Also== | ||
| Line 33: | Line 34: | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
[[Category: | [[Category: Bacillus coli migula 1895]] | ||
[[Category: Cramer, W A]] | [[Category: Cramer, W A]] | ||
[[Category: Sharma, O]] | [[Category: Sharma, O]] | ||
Revision as of 18:45, 7 February 2016
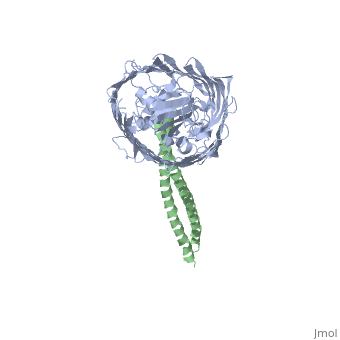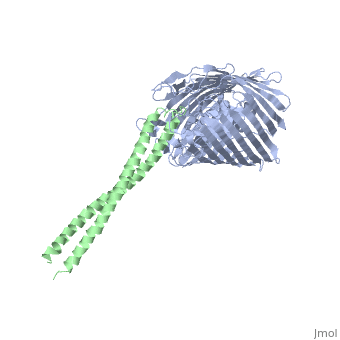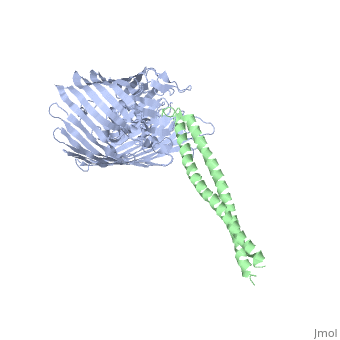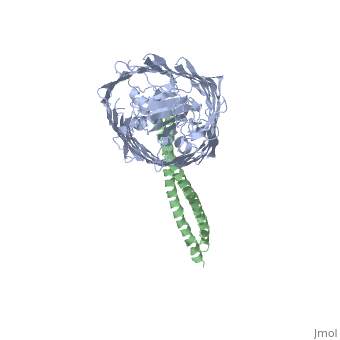
Structure of the complex between BtuB and Colicin E2 receptor binding domainStructure of the complex between BtuB and Colicin E2 receptor binding domain
Structural highlights
Function[BTUB_ECOLI] Involved in the active translocation of vitamin B12 (cyanocobalamin) across the outer membrane to the periplasmic space. It derives its energy for transport by interacting with the trans-periplasmic membrane protein TonB. Is also a receptor for bacteriophages BF23 and C1, and for A and E colicins.[HAMAP-Rule:MF_01531] [CEA2_ECOLX] This plasmid-coded bactericidal protein is an endonuclease active on both single- and double-stranded DNA but with undefined specificity. Colicins are polypeptide toxins produced by and active against E.coli and closely related bacteria. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe crystal structure of the complex of the BtuB receptor and the 135-residue coiled-coil receptor-binding R-domain of colicin E3 (E3R135) suggested a novel mechanism for import of colicin proteins across the outer membrane. It was proposed that one function of the R-domain, which extends along the outer membrane surface, is to recruit an additional outer membrane protein(s) to form a translocon for passage colicin activity domain. A 3.5-A crystal structure of the complex of E2R135 and BtuB (E2R135-BtuB) was obtained, which revealed E2R135 bound to BtuB in an oblique orientation identical to that previously found for E3R135. The only significant difference between the two structures was that the bound coiled-coil R-domain of colicin E2, compared with that of colicin E3, was extended by two and five residues at the N and C termini, respectively. There was no detectable displacement of the BtuB plug domain in either structure, implying that colicin is not imported through the outer membrane by BtuB alone. It was concluded that the oblique orientation of the R-domain of the nuclease E colicins has a function in the recruitment of another member(s) of an outer membrane translocon. Screening of porin knock-out mutants showed that either OmpF or OmpC can function in such a translocon. Arg(452) at the R/C-domain interface in colicin E2 was found have an essential role at a putative site of protease cleavage, which would liberate the C-terminal activity domain for passage through the outer membrane translocon. Structure of the complex of the colicin E2 R-domain and its BtuB receptor. The outer membrane colicin translocon.,Sharma O, Yamashita E, Zhalnina MV, Zakharov SD, Datsenko KA, Wanner BL, Cramer WA J Biol Chem. 2007 Aug 10;282(32):23163-70. Epub 2007 Jun 4. PMID:17548346[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||