1a32: Difference between revisions
No edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
|DOMAIN= | |||
|RELATEDENTRY= | |||
|RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1a32 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1a32 OCA], [http://www.ebi.ac.uk/pdbsum/1a32 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1a32 RCSB]</span> | |||
}} | }} | ||
| Line 33: | Line 36: | ||
[[Category: x-ray crystallography]] | [[Category: x-ray crystallography]] | ||
''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 18:32:08 2008'' | ||
Revision as of 18:32, 30 March 2008
| |||||||
| , resolution 2.10Å | |||||||
|---|---|---|---|---|---|---|---|
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
RIBOSOMAL PROTEIN S15 FROM BACILLUS STEAROTHERMOPHILUS
OverviewOverview
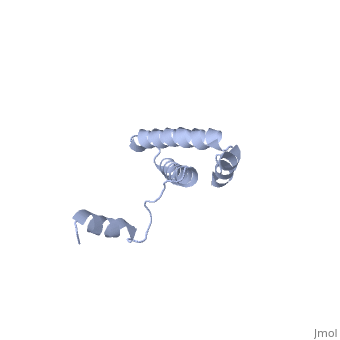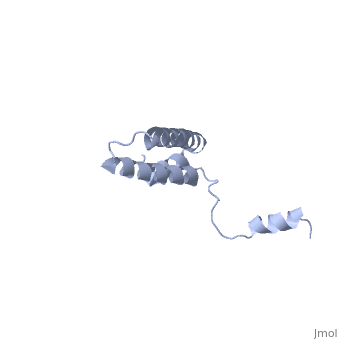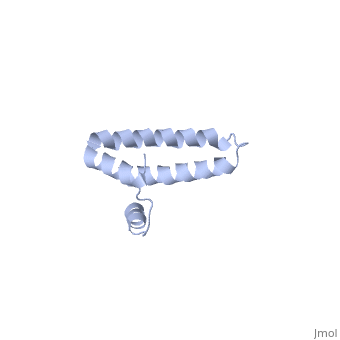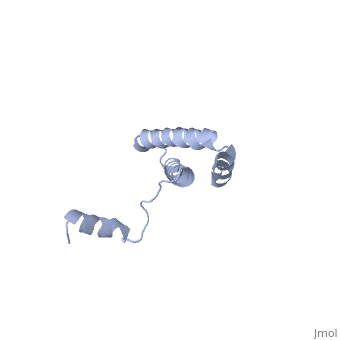
BACKGROUND: Ribosomal protein S15 is a primary RNA-binding protein that binds to the central domain of 16S rRNA. S15 also regulates its own synthesis by binding to its own mRNA. The binding sites for S15 on both mRNA and rRNA have been narrowed down to less than a hundred nucleotides each, making the protein an attractive candidate for the study of protein-RNA interactions. RESULTS: The crystal structure of S15 from Bacillus stearothermophilus has been solved to 2.1 A resolution. The structure consists of four alpha helices. Three of these helices form the core of the protein, while the N-terminal helix protrudes out from the body of the molecule to make contacts with a neighboring molecule in the crystal lattice. S15 contains a large conserved patch of basic residues which could provide a site for binding 16S rRNA. CONCLUSIONS: The conformation of the N-terminal alpha helix is quite different from that reported in a recent NMR structure of S15 from Thermus thermophilus. The intermolecular contacts that this alpha helix makes with a neighboring molecule in the crystal, however, closely resemble the intramolecular contacts that occur in the NMR structure. This conformational variability of the N-terminal helix has implications for the range of possible S15-RNA interactions. A large, conserved basic patch at one end of S15 and a cluster of conserved but exposed aromatic residues at the other end provide two possible RNA-binding sites on S15.
About this StructureAbout this Structure
1A32 is a Single protein structure of sequence from Geobacillus stearothermophilus. Full crystallographic information is available from OCA.
ReferenceReference
Conformational variability of the N-terminal helix in the structure of ribosomal protein S15., Clemons WM Jr, Davies C, White SW, Ramakrishnan V, Structure. 1998 Apr 15;6(4):429-38. PMID:9562554
Page seeded by OCA on Sun Mar 30 18:32:08 2008