1a0a: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
|PDB= 1a0a |SIZE=350|CAPTION= <scene name='initialview01'>1a0a</scene>, resolution 2.800Å | |PDB= 1a0a |SIZE=350|CAPTION= <scene name='initialview01'>1a0a</scene>, resolution 2.800Å | ||
|SITE= | |SITE= | ||
|LIGAND= | |LIGAND= <scene name='pdbligand=DA:2'-DEOXYADENOSINE-5'-MONOPHOSPHATE'>DA</scene>, <scene name='pdbligand=DC:2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE'>DC</scene>, <scene name='pdbligand=DG:2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE'>DG</scene>, <scene name='pdbligand=DT:THYMIDINE-5'-MONOPHOSPHATE'>DT</scene> | ||
|ACTIVITY= | |ACTIVITY= | ||
|GENE= | |GENE= | ||
|DOMAIN= | |||
|RELATEDENTRY= | |||
|RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1a0a FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1a0a OCA], [http://www.ebi.ac.uk/pdbsum/1a0a PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=1a0a RCSB]</span> | |||
}} | }} | ||
| Line 34: | Line 37: | ||
[[Category: transcription factor]] | [[Category: transcription factor]] | ||
''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sun Mar 30 18:30:25 2008'' | ||
Revision as of 18:30, 30 March 2008
| |||||||
| , resolution 2.800Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , , | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
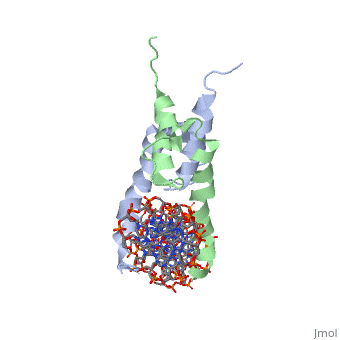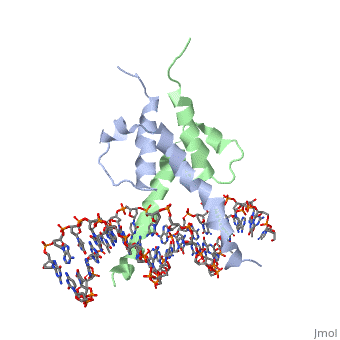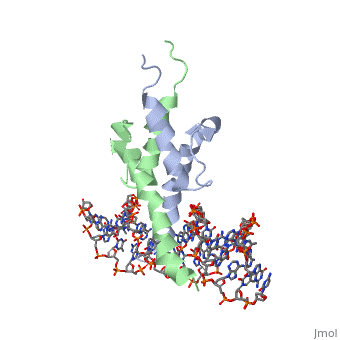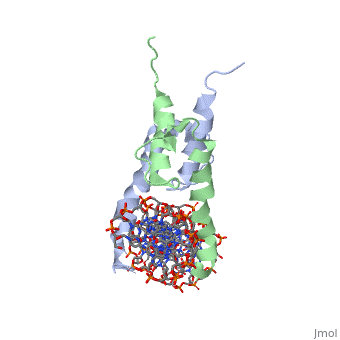
PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX
OverviewOverview
The crystal structure of a DNA-binding domain of PHO4 complexed with DNA at 2.8 A resolution revealed that the domain folds into a basic-helix-loop-helix (bHLH) motif with a long but compact loop that contains a short alpha-helical segment. This helical structure positions a tryptophan residue into an aromatic cluster so as to make the loop compact. PHO4 binds to DNA as a homodimer with direct reading of both the core E-box sequence CACGTG and its 3'-flanking bases. The 3'-flanking bases GG are recognized by Arg2 and His5. The residues involved in the E-box recognition are His5, Glu9 and Arg13, as already reported for bHLH/Zip proteins MAX and USF, and are different from those recognized by bHLH proteins MyoD and E47, although PHO4 is a bHLH protein.
About this StructureAbout this Structure
1A0A is a Single protein structure of sequence from Saccharomyces cerevisiae. Full crystallographic information is available from OCA.
ReferenceReference
Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition., Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T, EMBO J. 1997 Aug 1;16(15):4689-97. PMID:9303313
Page seeded by OCA on Sun Mar 30 18:30:25 2008