2exs: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:2exs.gif|left|200px]] | [[Image:2exs.gif|left|200px]] | ||
'''TRAP3 (engineered TRAP)''' | {{Structure | ||
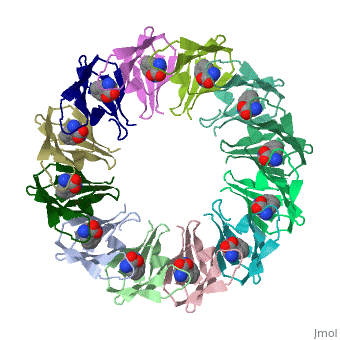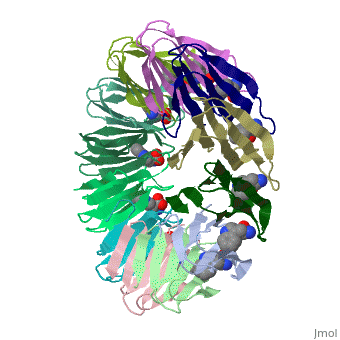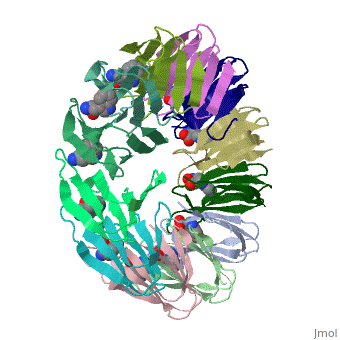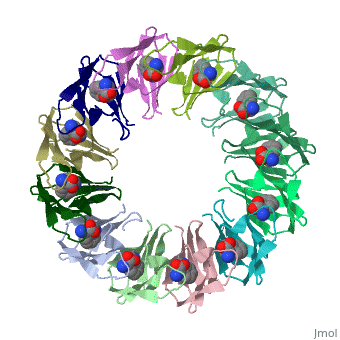
|PDB= 2exs |SIZE=350|CAPTION= <scene name='initialview01'>2exs</scene>, resolution 2.00Å | |||
|SITE= | |||
|LIGAND= <scene name='pdbligand=TRP:TRYPTOPHAN'>TRP</scene> and <scene name='pdbligand=TRP:TRYPTOPHAN'>TRP</scene> | |||
|ACTIVITY= | |||
|GENE= mtrB ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=1422 Geobacillus stearothermophilus]) | |||
}} | |||
'''TRAP3 (engineered TRAP)''' | |||
==Overview== | ==Overview== | ||
| Line 7: | Line 16: | ||
==About this Structure== | ==About this Structure== | ||
2EXS is a [ | 2EXS is a [[Single protein]] structure of sequence from [http://en.wikipedia.org/wiki/Geobacillus_stearothermophilus Geobacillus stearothermophilus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2EXS OCA]. | ||
==Reference== | ==Reference== | ||
Rounding up: Engineering 12-membered rings from the cyclic 11-mer TRAP., Heddle JG, Yokoyama T, Yamashita I, Park SY, Tame JR, Structure. 2006 May;14(5):925-33. PMID:[http:// | Rounding up: Engineering 12-membered rings from the cyclic 11-mer TRAP., Heddle JG, Yokoyama T, Yamashita I, Park SY, Tame JR, Structure. 2006 May;14(5):925-33. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/16698553 16698553] | ||
[[Category: Geobacillus stearothermophilus]] | [[Category: Geobacillus stearothermophilus]] | ||
[[Category: Single protein]] | [[Category: Single protein]] | ||
| Line 24: | Line 33: | ||
[[Category: ring protein]] | [[Category: ring protein]] | ||
''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Mar 20 16:45:39 2008'' | ||
Revision as of 17:45, 20 March 2008
| |||||||
| , resolution 2.00Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | and | ||||||
| Gene: | mtrB (Geobacillus stearothermophilus) | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
TRAP3 (engineered TRAP)
OverviewOverview
The protein TRAP (trp RNA binding attenuation protein) forms a highly thermostable ring-shaped 11-mer. By linking in tandem two, three, or four DNA sequences encoding TRAP monomers, we have engineered new rings that consist of 12 TRAP subunits and bind 12 ligand molecules. The hydrogen bonding pattern and buried surface area within and between subunits are essentially identical between the 11-mer and 12-mer crystal structures. Why do the artificial proteins choose to make single 12-mer rings? The 12-mer rings are highly sterically strained by their peptide linkers and far from thermostable. That proteins choose to adopt a strained conformation of few subunits rather than an unstrained one with 11 subunits demonstrates the importance of entropic factors in controlling protein-protein interactions in general.
About this StructureAbout this Structure
2EXS is a Single protein structure of sequence from Geobacillus stearothermophilus. Full crystallographic information is available from OCA.
ReferenceReference
Rounding up: Engineering 12-membered rings from the cyclic 11-mer TRAP., Heddle JG, Yokoyama T, Yamashita I, Park SY, Tame JR, Structure. 2006 May;14(5):925-33. PMID:16698553
Page seeded by OCA on Thu Mar 20 16:45:39 2008