1uug: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:1uug.jpg|left|200px]] | [[Image:1uug.jpg|left|200px]] | ||
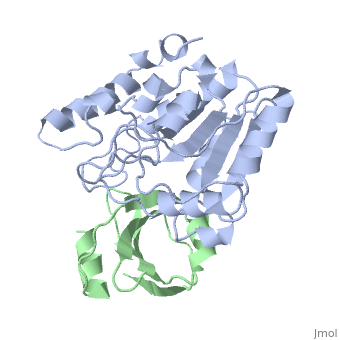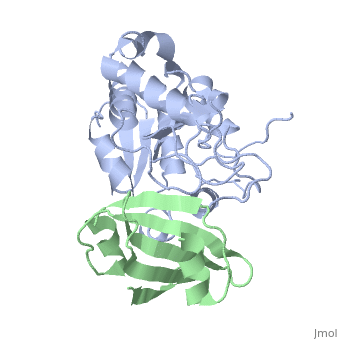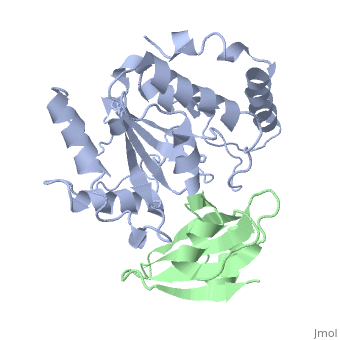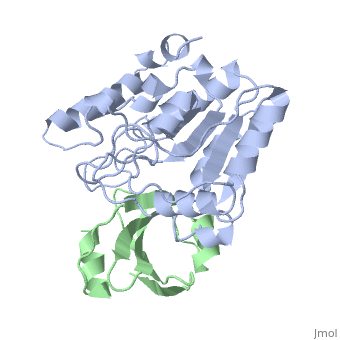
'''ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI''' | {{Structure | ||
|PDB= 1uug |SIZE=350|CAPTION= <scene name='initialview01'>1uug</scene>, resolution 2.4Å | |||
|SITE= | |||
|LIGAND= | |||
|ACTIVITY= [http://en.wikipedia.org/wiki/Uridine_nucleosidase Uridine nucleosidase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.2.2.3 3.2.2.3] | |||
|GENE= UNG ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=562 Escherichia coli]) | |||
}} | |||
'''ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI''' | |||
==Overview== | ==Overview== | ||
| Line 7: | Line 16: | ||
==About this Structure== | ==About this Structure== | ||
1UUG is a [ | 1UUG is a [[Protein complex]] structure of sequences from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli] and [http://en.wikipedia.org/wiki/Phage_pbs1 Phage pbs1]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1UUG OCA]. | ||
==Reference== | ==Reference== | ||
Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase., Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA, J Mol Biol. 1999 Mar 26;287(2):331-46. PMID:[http:// | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase., Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA, J Mol Biol. 1999 Mar 26;287(2):331-46. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/10080896 10080896] | ||
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
[[Category: Phage pbs1]] | [[Category: Phage pbs1]] | ||
| Line 23: | Line 32: | ||
[[Category: protein mimicry of dna]] | [[Category: protein mimicry of dna]] | ||
''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Mar 20 14:36:58 2008'' | ||
Revision as of 15:37, 20 March 2008
| |||||||
| , resolution 2.4Å | |||||||
|---|---|---|---|---|---|---|---|
| Gene: | UNG (Escherichia coli) | ||||||
| Activity: | Uridine nucleosidase, with EC number 3.2.2.3 | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI
OverviewOverview
Uracil-DNA glycosylase (UDG), which is a critical enzyme in DNA base-excision repair that recognizes and removes uracil from DNA, is specifically and irreversably inhibited by the thermostable uracil-DNA glycosylase inhibitor protein (Ugi). A paradox for the highly specific Ugi inhibition of UDG is how Ugi can successfully mimic DNA backbone interactions for UDG without resulting in significant cross-reactivity with numerous other enzymes that possess DNA backbone binding affinity. High-resolution X-ray crystal structures of Ugi both free and in complex with wild-type and the functionally defective His187Asp mutant Escherichia coli UDGs reveal the detailed molecular basis for duplex DNA backbone mimicry by Ugi. The overall shape and charge distribution of Ugi most closely resembles a midpoint in a trajectory between B-form DNA and the kinked DNA observed in UDG:DNA product complexes. Thus, Ugi targets the mechanism of uracil flipping by UDG and appears to be a transition-state mimic for UDG-flipping of uracil nucleotides from DNA. Essentially all the exquisite shape, electrostatic and hydrophobic complementarity for the high-affinity UDG-Ugi interaction is pre-existing, except for a key flip of the Ugi Gln19 carbonyl group and Glu20 side-chain, which is triggered by the formation of the complex. Conformational changes between unbound Ugi and Ugi complexed with UDG involve the beta-zipper structural motif, which we have named for the reversible pairing observed between intramolecular beta-strands. A similar beta-zipper is observed in the conversion between the open and closed forms of UDG. The combination of extremely high levels of pre-existing structural complementarity to DNA binding features specific to UDG with key local conformational changes in Ugi resolves the UDG-Ugi paradox and suggests a potentially general structural solution to the formation of very high affinity DNA enzyme-inhibitor complexes that avoid cross- reactivity.
About this StructureAbout this Structure
1UUG is a Protein complex structure of sequences from Escherichia coli and Phage pbs1. Full crystallographic information is available from OCA.
ReferenceReference
Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase., Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA, J Mol Biol. 1999 Mar 26;287(2):331-46. PMID:10080896
Page seeded by OCA on Thu Mar 20 14:36:58 2008