3pih: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
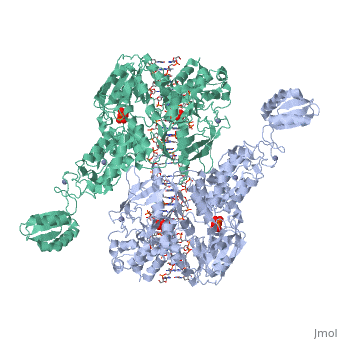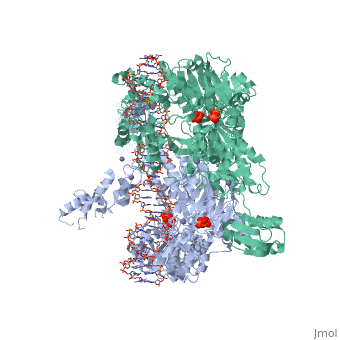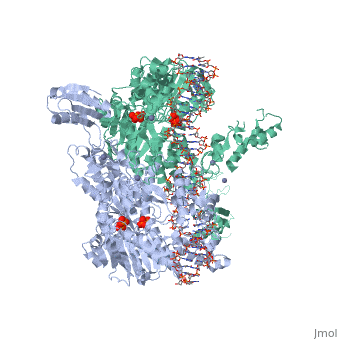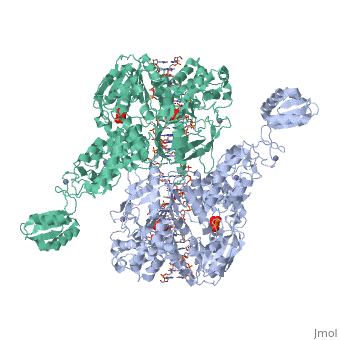
==T. maritima UvrA in complex with fluorescein-modified DNA== | |||
=== | <StructureSection load='3pih' size='340' side='right' caption='[[3pih]], [[Resolution|resolution]] 2.90Å' scene=''> | ||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[3pih]] is a 2 chain structure with sequence from [http://en.wikipedia.org/wiki/Thermotoga_maritima Thermotoga maritima]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3PIH OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3PIH FirstGlance]. <br> | |||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=PPV:PYROPHOSPHATE'>PPV</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | |||
<tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">uvrA, TM_0480 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=2336 Thermotoga maritima])</td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3pih FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3pih OCA], [http://www.rcsb.org/pdb/explore.do?structureId=3pih RCSB], [http://www.ebi.ac.uk/pdbsum/3pih PDBsum]</span></td></tr> | |||
</table> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
One of the primary pathways for removal of DNA damage is nucleotide excision repair (NER). In bacteria, the UvrA protein is the component of NER that locates the lesion. A notable feature of NER is its ability to act on many DNA modifications that vary in chemical structure. So far, the mechanism underlying this broad specificity has been unclear. Here, we report the first crystal structure of a UvrA protein in complex with a chemically modified oligonucleotide. The structure shows that the UvrA dimer does not contact the site of lesion directly, but rather binds the DNA regions on both sides of the modification. The DNA region harboring the modification is deformed, with the double helix bent and unwound. UvrA uses damage-induced deformations of the DNA and a less rigid structure of the modified double helix for indirect readout of the lesion. | |||
Structure of UvrA nucleotide excision repair protein in complex with modified DNA.,Jaciuk M, Nowak E, Skowronek K, Tanska A, Nowotny M Nat Struct Mol Biol. 2011 Feb;18(2):191-7. Epub 2011 Jan 16. PMID:21240268<ref>PMID:21240268</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
==See Also== | ==See Also== | ||
*[[UvrABC|UvrABC]] | *[[UvrABC|UvrABC]] | ||
== References == | |||
== | <references/> | ||
__TOC__ | |||
</StructureSection> | |||
[[Category: Thermotoga maritima]] | [[Category: Thermotoga maritima]] | ||
[[Category: Jaciuk, M | [[Category: Jaciuk, M]] | ||
[[Category: Nowak, E | [[Category: Nowak, E]] | ||
[[Category: Nowotny, M | [[Category: Nowotny, M]] | ||
[[Category: Abc atpase]] | [[Category: Abc atpase]] | ||
[[Category: Dna repair]] | [[Category: Dna repair]] | ||
Revision as of 10:07, 19 December 2014
T. maritima UvrA in complex with fluorescein-modified DNAT. maritima UvrA in complex with fluorescein-modified DNA
Structural highlights
Publication Abstract from PubMedOne of the primary pathways for removal of DNA damage is nucleotide excision repair (NER). In bacteria, the UvrA protein is the component of NER that locates the lesion. A notable feature of NER is its ability to act on many DNA modifications that vary in chemical structure. So far, the mechanism underlying this broad specificity has been unclear. Here, we report the first crystal structure of a UvrA protein in complex with a chemically modified oligonucleotide. The structure shows that the UvrA dimer does not contact the site of lesion directly, but rather binds the DNA regions on both sides of the modification. The DNA region harboring the modification is deformed, with the double helix bent and unwound. UvrA uses damage-induced deformations of the DNA and a less rigid structure of the modified double helix for indirect readout of the lesion. Structure of UvrA nucleotide excision repair protein in complex with modified DNA.,Jaciuk M, Nowak E, Skowronek K, Tanska A, Nowotny M Nat Struct Mol Biol. 2011 Feb;18(2):191-7. Epub 2011 Jan 16. PMID:21240268[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||