Pheromone binding protein: Difference between revisions
Nurit Eliash (talk | contribs) No edit summary |
Nurit Eliash (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
==Introduction== | ==Introduction== | ||
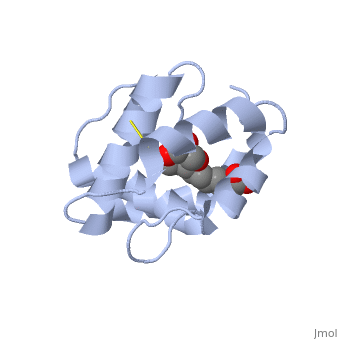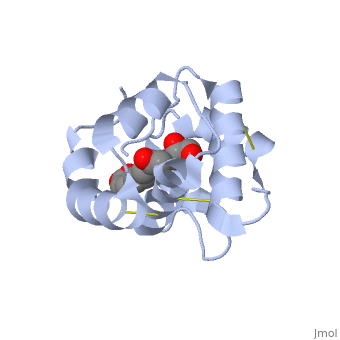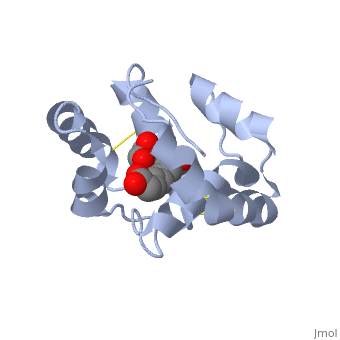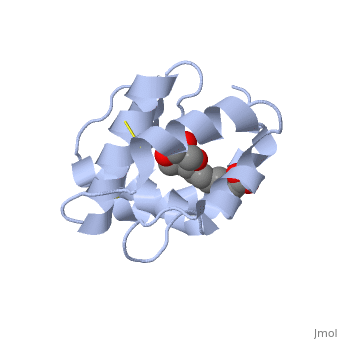
<StructureSection load='3bfa' size='340' side='right' caption='Pheromone binding protein of ''Apis mellifera'' scene=''> | <StructureSection load='3bfa' size='340' side='right' caption='Pheromone binding protein of ''Apis mellifera'' scene=''> | ||
Pheromone binding proteins are soluble proteins | Pheromone binding proteins [http://en.wikipedia.org/wiki/Pheromone_binding_protein (PBP)] are type of Odorant binding proteins [http://en.wikipedia.org/wiki/Odorant-binding_protein (OBP)] - soluble proteins mediating the early stages of volatiles detection in both insects and vertebrates | ||
As a model protein of this family we will further describe the well investigated Pheromone binding protein of the honey bee. | <ref>DOI:10.3389/fphys.2014.00320</ref>. | ||
The volatiles (pheromones and other small hydrophobic molecules) are solubilized by the OBPs and activate the chemoreceptors. | |||
As a model protein of this family we will further describe the well investigated Pheromone binding protein of the honey bee, ASP1. | |||
== Pheromone-binding protein ASP1 == | |||
Chemical communication is crucial in social insects, where a complicated and delicate system of signals must be maintained in order to preserve the fragile equilibrium between the queen and the workers. | Chemical communication is crucial in social insects, where a complicated and delicate system of signals must be maintained in order to preserve the fragile equilibrium between the queen and the workers. | ||
In the hive of the honey bee [http://en.wikipedia.org/wiki/Apis_mellifera ''Apis mellifera''] this equilibrium exists partially due to the extraction of blend of substances called queen mandibular pheromone [http://en.wikipedia.org/wiki/Honey_bee_pheromones#Queen_mandibular_pheromone (QMP)], by the queen. The three major component of the QMP blend are: 9-keto-2(E)-decenoic acid (9-ODA) and 9-hydroxy-2(E)-decenoic acid (9-HDA R-(−) or S-(+)). | In the hive of the honey bee [http://en.wikipedia.org/wiki/Apis_mellifera ''Apis mellifera''] this equilibrium exists partially due to the extraction of blend of substances called queen mandibular pheromone [http://en.wikipedia.org/wiki/Honey_bee_pheromones#Queen_mandibular_pheromone (QMP)], by the queen <ref>Winston, M.L., 1987. The Biology of the Honey Bee. Harvard University Press, Cambridge, MA.</ref>. The three major component of the QMP blend are: 9-keto-2(E)-decenoic acid (9-ODA) and 9-hydroxy-2(E)-decenoic acid (9-HDA R-(−) or S-(+)). | ||
Pheromone-binding protein [http://www.uniprot.org/uniprot/Q9U9J6/ ASP] of the honeybee [http://en.wikipedia.org/wiki/Apis_mellifera ''Apis mellifera''] L. (Hymenoptera: Apidea) was first isolated and characterized by Danty ''et al''. (1998)<ref>DOI:10.1016/j.jmb.2008.04.048</ref> from the bee antennae. | |||
Pheromone-binding protein [http://www.uniprot.org/uniprot/Q9U9J6/ ASP] of the honeybee [http://en.wikipedia.org/wiki/Apis_mellifera ''Apis mellifera''] L. (Hymenoptera: Apidea) was first isolated and characterized by Danty ''et al''. (1998)<ref>DOI:10.1016/j.jmb.2008.04.048</ref> from the bee antennae. | |||
== Structure == | |||
The protein is composed of 144 amino acids, which forms 6 alpha helices. Three <scene name='60/609542/Disulfide_bonds/1'>3 disulfide bonds</scene> tied four helices: disulfide 20–51 between H1 and H3, 47– 98 between H3 and H6, and 107–89 between H6 and H5. | |||
== Interaction with the ligand 9-ODA== | |||
<scene name='60/609542/9-oda/3'>9-ODA</scene> | |||
The carboxyl end of the component 9-ODA points towards the solvent, and has no any interaction with residues of the protein. The residues in the binding site are <scene name='60/609542/Binding_site/3'>hydrophobic</scene> | |||
, and the connection between 9-ODA and ASP1 involve hydrogen bonds. | |||
---- | ---- | ||
<scene name='60/609542/Glycerol/2'>Glycerol</scene> | <scene name='60/609542/Glycerol/2'>Glycerol</scene> | ||
or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
Revision as of 14:27, 22 November 2014
IntroductionIntroduction
Pheromone binding proteins (PBP) are type of Odorant binding proteins (OBP) - soluble proteins mediating the early stages of volatiles detection in both insects and vertebrates [1]. The volatiles (pheromones and other small hydrophobic molecules) are solubilized by the OBPs and activate the chemoreceptors. As a model protein of this family we will further describe the well investigated Pheromone binding protein of the honey bee, ASP1. Pheromone-binding protein ASP1Chemical communication is crucial in social insects, where a complicated and delicate system of signals must be maintained in order to preserve the fragile equilibrium between the queen and the workers. In the hive of the honey bee Apis mellifera this equilibrium exists partially due to the extraction of blend of substances called queen mandibular pheromone (QMP), by the queen [2]. The three major component of the QMP blend are: 9-keto-2(E)-decenoic acid (9-ODA) and 9-hydroxy-2(E)-decenoic acid (9-HDA R-(−) or S-(+)). Pheromone-binding protein ASP of the honeybee Apis mellifera L. (Hymenoptera: Apidea) was first isolated and characterized by Danty et al. (1998)[3] from the bee antennae. Pheromone-binding protein ASP of the honeybee Apis mellifera L. (Hymenoptera: Apidea) was first isolated and characterized by Danty et al. (1998)[4] from the bee antennae. StructureThe protein is composed of 144 amino acids, which forms 6 alpha helices. Three tied four helices: disulfide 20–51 between H1 and H3, 47– 98 between H3 and H6, and 107–89 between H6 and H5. Interaction with the ligand 9-ODA
The carboxyl end of the component 9-ODA points towards the solvent, and has no any interaction with residues of the protein. The residues in the binding site are , and the connection between 9-ODA and ASP1 involve hydrogen bonds.
or to the article describing Jmol [5] to the rescue. StructureThe protein has a 1 alpha chain, that can be seen in pink.
FunctionThe protein ligand, 9-ODA File:9-ODA.jpg Structural highlightsThis is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
|
| ||||||||||
ReferencesReferences
- ↑ Pelosi P, Iovinella I, Felicioli A, Dani FR. Soluble proteins of chemical communication: an overview across arthropods. Front Physiol. 2014 Aug 27;5:320. doi: 10.3389/fphys.2014.00320. eCollection, 2014. PMID:25221516 doi:http://dx.doi.org/10.3389/fphys.2014.00320
- ↑ Winston, M.L., 1987. The Biology of the Honey Bee. Harvard University Press, Cambridge, MA.
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Tegoni M, Cambillau C. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J Mol Biol. 2008 Jun 27;380(1):158-69. Epub 2008 Apr 27. PMID:18508083 doi:10.1016/j.jmb.2008.04.048
- ↑ Pesenti ME, Spinelli S, Bezirard V, Briand L, Pernollet JC, Tegoni M, Cambillau C. Structural basis of the honey bee PBP pheromone and pH-induced conformational change. J Mol Biol. 2008 Jun 27;380(1):158-69. Epub 2008 Apr 27. PMID:18508083 doi:10.1016/j.jmb.2008.04.048
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644