1xbn: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
==Crystal structure of a bacterial nitric oxide sensor: an ortholog of mammalian soluble guanylate cyclase heme domain== | |||
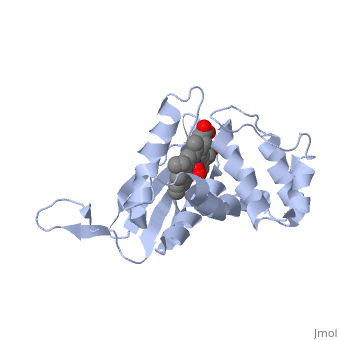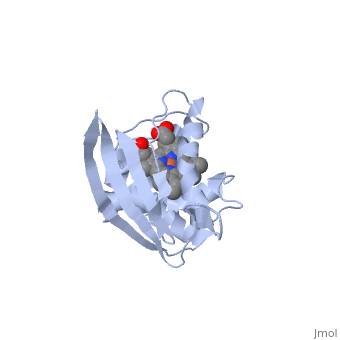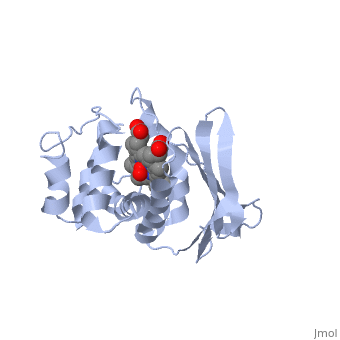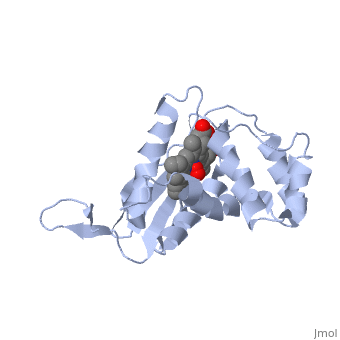
<StructureSection load='1xbn' size='340' side='right' caption='[[1xbn]], [[Resolution|resolution]] 2.50Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1xbn]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Thermoanaerobacter_tengcongensis_mb4 Thermoanaerobacter tengcongensis mb4]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1XBN OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1XBN FirstGlance]. <br> | |||
</td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=HEM:PROTOPORPHYRIN+IX+CONTAINING+FE'>HEM</scene>, <scene name='pdbligand=OXY:OXYGEN+MOLECULE'>OXY</scene><br> | |||
<tr><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">Tar4 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=273068 Thermoanaerobacter tengcongensis MB4])</td></tr> | |||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1xbn FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1xbn OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1xbn RCSB], [http://www.ebi.ac.uk/pdbsum/1xbn PDBsum]</span></td></tr> | |||
<table> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/xb/1xbn_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Nitric oxide (NO) is extremely toxic to Clostridium botulinum, but its molecular targets are unknown. Here, we identify a heme protein sensor (SONO) that displays femtomolar affinity for NO. The crystal structure of the SONO heme domain reveals a previously undescribed fold and a strategically placed tyrosine residue that modulates heme-nitrosyl coordination. Furthermore, the domain architecture of a SONO ortholog cloned from Chlamydomonas reinhardtii indicates that NO signaling through cyclic guanosine monophosphate arose before the origin of multicellular eukaryotes. Our findings have broad implications for understanding bacterial responses to NO, as well as for the activation of mammalian NO-sensitive guanylyl cyclase. | |||
Femtomolar sensitivity of a NO sensor from Clostridium botulinum.,Nioche P, Berka V, Vipond J, Minton N, Tsai AL, Raman CS Science. 2004 Nov 26;306(5701):1550-3. Epub 2004 Oct 7. PMID:15472039<ref>PMID:15472039</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
==See Also== | ==See Also== | ||
*[[Chemotaxis protein|Chemotaxis protein]] | *[[Chemotaxis protein|Chemotaxis protein]] | ||
*[[Methyl-accepting chemotaxis protein|Methyl-accepting chemotaxis protein]] | *[[Methyl-accepting chemotaxis protein|Methyl-accepting chemotaxis protein]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Thermoanaerobacter tengcongensis mb4]] | [[Category: Thermoanaerobacter tengcongensis mb4]] | ||
[[Category: Nioche, P.]] | [[Category: Nioche, P.]] | ||
Revision as of 23:15, 29 September 2014
Crystal structure of a bacterial nitric oxide sensor: an ortholog of mammalian soluble guanylate cyclase heme domainCrystal structure of a bacterial nitric oxide sensor: an ortholog of mammalian soluble guanylate cyclase heme domain
Structural highlights
Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedNitric oxide (NO) is extremely toxic to Clostridium botulinum, but its molecular targets are unknown. Here, we identify a heme protein sensor (SONO) that displays femtomolar affinity for NO. The crystal structure of the SONO heme domain reveals a previously undescribed fold and a strategically placed tyrosine residue that modulates heme-nitrosyl coordination. Furthermore, the domain architecture of a SONO ortholog cloned from Chlamydomonas reinhardtii indicates that NO signaling through cyclic guanosine monophosphate arose before the origin of multicellular eukaryotes. Our findings have broad implications for understanding bacterial responses to NO, as well as for the activation of mammalian NO-sensitive guanylyl cyclase. Femtomolar sensitivity of a NO sensor from Clostridium botulinum.,Nioche P, Berka V, Vipond J, Minton N, Tsai AL, Raman CS Science. 2004 Nov 26;306(5701):1550-3. Epub 2004 Oct 7. PMID:15472039[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences |
| ||||||||||||||||||