1aoi: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
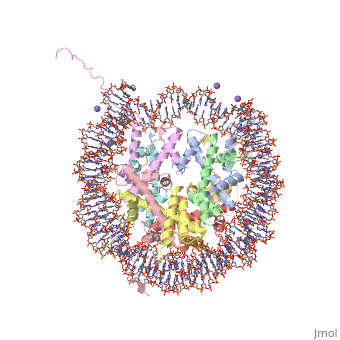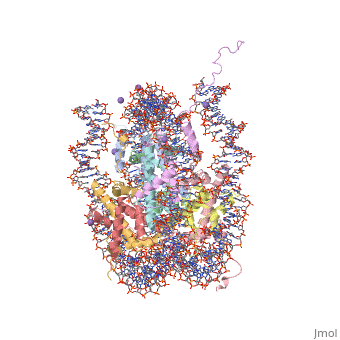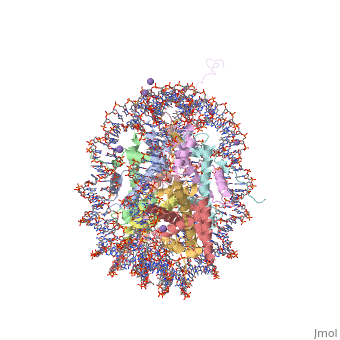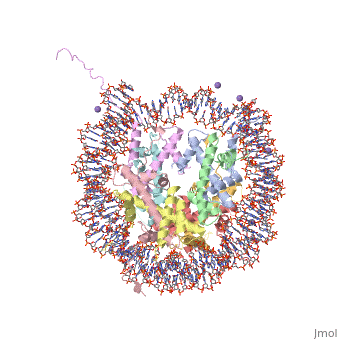
==COMPLEX BETWEEN NUCLEOSOME CORE PARTICLE (H3,H4,H2A,H2B) AND 146 BP LONG DNA FRAGMENT== | |||
<StructureSection load='1aoi' size='340' side='right' caption='[[1aoi]], [[Resolution|resolution]] 2.80Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1aoi]] is a 10 chain structure with sequence from [http://en.wikipedia.org/wiki/Xenopus_laevis Xenopus laevis]. The July 2000 RCSB PDB [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Nucleosome'' by David S. Goodsell is [http://dx.doi.org/10.2210/rcsb_pdb/mom_2000_7 10.2210/rcsb_pdb/mom_2000_7]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1AOI OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1AOI FirstGlance]. <br> | |||
</td></tr><tr><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=MN:MANGANESE+(II)+ION'>MN</scene><br> | |||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1aoi FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1aoi OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1aoi RCSB], [http://www.ebi.ac.uk/pdbsum/1aoi PDBsum]</span></td></tr> | |||
<table> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ao/1aoi_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
The X-ray crystal structure of the nucleosome core particle of chromatin shows in atomic detail how the histone protein octamer is assembled and how 146 base pairs of DNA are organized into a superhelix around it. Both histone/histone and histone/DNA interactions depend on the histone fold domains and additional, well ordered structure elements extending from this motif. Histone amino-terminal tails pass over and between the gyres of the DNA superhelix to contact neighbouring particles. The lack of uniformity between multiple histone/DNA-binding sites causes the DNA to deviate from ideal superhelix geometry. | |||
Crystal structure of the nucleosome core particle at 2.8 A resolution.,Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ Nature. 1997 Sep 18;389(6648):251-60. PMID:9305837<ref>PMID:9305837</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
==See Also== | ==See Also== | ||
| Line 11: | Line 30: | ||
*[[User:Eric Martz/Nucleosomes|User:Eric Martz/Nucleosomes]] | *[[User:Eric Martz/Nucleosomes|User:Eric Martz/Nucleosomes]] | ||
*[[User:Luca Toldo|User:Luca Toldo]] | *[[User:Luca Toldo|User:Luca Toldo]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Nucleosome]] | [[Category: Nucleosome]] | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
Revision as of 20:20, 29 September 2014
COMPLEX BETWEEN NUCLEOSOME CORE PARTICLE (H3,H4,H2A,H2B) AND 146 BP LONG DNA FRAGMENTCOMPLEX BETWEEN NUCLEOSOME CORE PARTICLE (H3,H4,H2A,H2B) AND 146 BP LONG DNA FRAGMENT
Structural highlights
Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe X-ray crystal structure of the nucleosome core particle of chromatin shows in atomic detail how the histone protein octamer is assembled and how 146 base pairs of DNA are organized into a superhelix around it. Both histone/histone and histone/DNA interactions depend on the histone fold domains and additional, well ordered structure elements extending from this motif. Histone amino-terminal tails pass over and between the gyres of the DNA superhelix to contact neighbouring particles. The lack of uniformity between multiple histone/DNA-binding sites causes the DNA to deviate from ideal superhelix geometry. Crystal structure of the nucleosome core particle at 2.8 A resolution.,Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ Nature. 1997 Sep 18;389(6648):251-60. PMID:9305837[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||