1st6: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
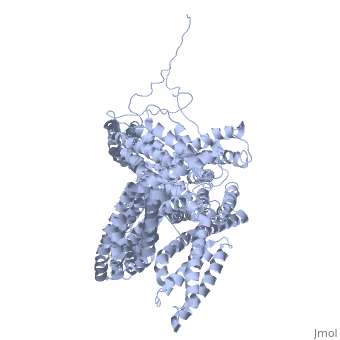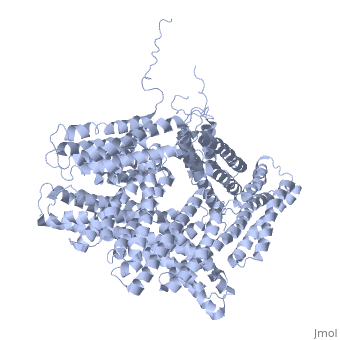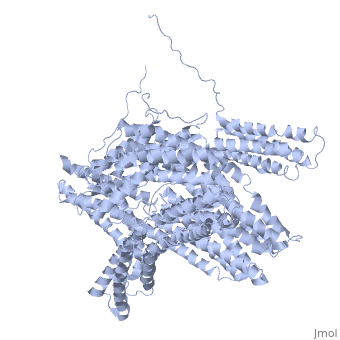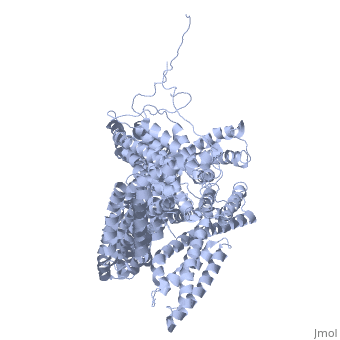
[[Image: | ==Crystal structure of a cytoskeletal protein== | ||
<StructureSection load='1st6' size='340' side='right' caption='[[1st6]], [[Resolution|resolution]] 3.10Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1st6]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Gallus_gallus Gallus gallus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1ST6 OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1ST6 FirstGlance]. <br> | |||
</td></tr><tr><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">VCL ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9031 Gallus gallus])</td></tr> | |||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1st6 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1st6 OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1st6 RCSB], [http://www.ebi.ac.uk/pdbsum/1st6 PDBsum]</span></td></tr> | |||
<table> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/st/1st6_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Vinculin is a highly conserved intracellular protein with a crucial role in the maintenance and regulation of cell adhesion and migration. In the cytosol, vinculin adopts a default autoinhibited conformation. On recruitment to cell-cell and cell-matrix adherens-type junctions, vinculin becomes activated and mediates various protein-protein interactions that regulate the links between F-actin and the cadherin and integrin families of cell-adhesion molecules. Here we describe the crystal structure of the full-length vinculin molecule (1,066 amino acids), which shows a five-domain autoinhibited conformation in which the carboxy-terminal tail domain is held pincer-like by the vinculin head, and ligand binding is regulated both sterically and allosterically. We show that conformational changes in the head, tail and proline-rich domains are linked structurally and thermodynamically, and propose a combinatorial pathway to activation that ensures that vinculin is activated only at sites of cell adhesion when two or more of its binding partners are brought into apposition. | |||
Structural basis for vinculin activation at sites of cell adhesion.,Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC Nature. 2004 Jul 29;430(6999):583-6. Epub 2004 Jun 13. PMID:15195105<ref>PMID:15195105</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
==See Also== | ==See Also== | ||
*[[Vinculin|Vinculin]] | *[[Vinculin|Vinculin]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Gallus gallus]] | [[Category: Gallus gallus]] | ||
[[Category: Bakolitsa, C.]] | [[Category: Bakolitsa, C.]] | ||
Revision as of 02:02, 29 September 2014
Crystal structure of a cytoskeletal proteinCrystal structure of a cytoskeletal protein
Structural highlights
Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedVinculin is a highly conserved intracellular protein with a crucial role in the maintenance and regulation of cell adhesion and migration. In the cytosol, vinculin adopts a default autoinhibited conformation. On recruitment to cell-cell and cell-matrix adherens-type junctions, vinculin becomes activated and mediates various protein-protein interactions that regulate the links between F-actin and the cadherin and integrin families of cell-adhesion molecules. Here we describe the crystal structure of the full-length vinculin molecule (1,066 amino acids), which shows a five-domain autoinhibited conformation in which the carboxy-terminal tail domain is held pincer-like by the vinculin head, and ligand binding is regulated both sterically and allosterically. We show that conformational changes in the head, tail and proline-rich domains are linked structurally and thermodynamically, and propose a combinatorial pathway to activation that ensures that vinculin is activated only at sites of cell adhesion when two or more of its binding partners are brought into apposition. Structural basis for vinculin activation at sites of cell adhesion.,Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC Nature. 2004 Jul 29;430(6999):583-6. Epub 2004 Jun 13. PMID:15195105[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||