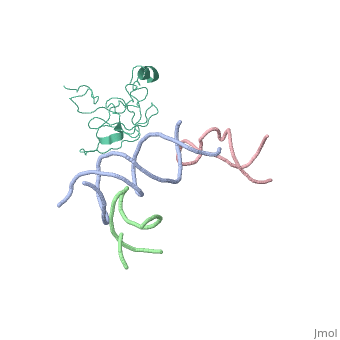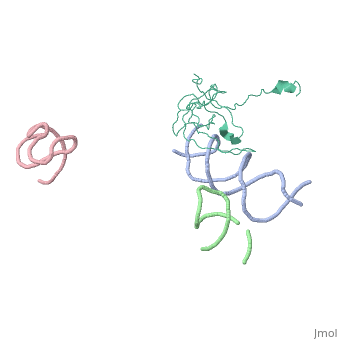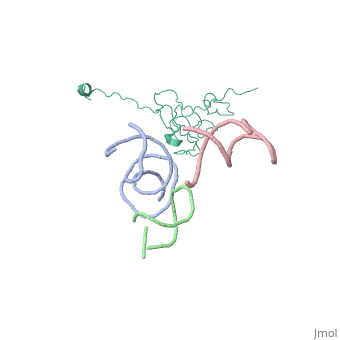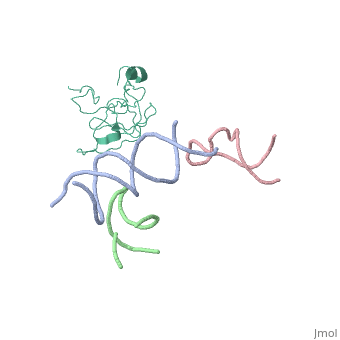1qzc: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image: | ==Coordinates of S12, SH44, LH69 and SRL separately fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome== | ||
<StructureSection load='1qzc' size='340' side='right' caption='[[1qzc]], [[Resolution|resolution]] 9.00Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1qzc]] is a 4 chain structure with sequence from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli] and [http://en.wikipedia.org/wiki/Thermus_thermophilus Thermus thermophilus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1QZC OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1QZC FirstGlance]. <br> | |||
</td></tr><tr><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1ffk|1ffk]], [[1ibm|1ibm]], [[1qza|1qza]], [[1qzb|1qzb]], [[1qzd|1qzd]], [[1r2w|1r2w]], [[1r2x|1r2x]]</td></tr> | |||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1qzc FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1qzc OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1qzc RCSB], [http://www.ebi.ac.uk/pdbsum/1qzc PDBsum]</span></td></tr> | |||
<table> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/qz/1qzc_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Aminoacyl-tRNAs (aa-tRNAs) are delivered to the ribosome as part of the ternary complex of aa-tRNA, elongation factor Tu (EF-Tu) and GTP. Here, we present a cryo-electron microscopy (cryo-EM) study, at a resolution of approximately 9 A, showing that during the incorporation of the aa-tRNA into the 70S ribosome of Escherichia coli, the flexibility of aa-tRNA allows the initial codon recognition and its accommodation into the ribosomal A site. In addition, a conformational change observed in the GTPase-associated center (GAC) of the ribosomal 50S subunit may provide the mechanism by which the ribosome promotes a relative movement of the aa-tRNA with respect to EF-Tu. This relative rearrangement seems to facilitate codon recognition by the incoming aa-tRNA, and to provide the codon-anticodon recognition-dependent signal for the GTPase activity of EF-Tu. From these new findings we propose a mechanism that can explain the sequence of events during the decoding of mRNA on the ribosome. | |||
Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy.,Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J Nat Struct Biol. 2003 Nov;10(11):899-906. Epub 2003 Oct 19. PMID:14566331<ref>PMID:14566331</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
==See Also== | ==See Also== | ||
*[[Ribosomal protein S12|Ribosomal protein S12]] | *[[Ribosomal protein S12|Ribosomal protein S12]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
[[Category: Thermus thermophilus]] | [[Category: Thermus thermophilus]] | ||
Revision as of 01:01, 29 September 2014
Coordinates of S12, SH44, LH69 and SRL separately fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosomeCoordinates of S12, SH44, LH69 and SRL separately fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome
Structural highlights
Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedAminoacyl-tRNAs (aa-tRNAs) are delivered to the ribosome as part of the ternary complex of aa-tRNA, elongation factor Tu (EF-Tu) and GTP. Here, we present a cryo-electron microscopy (cryo-EM) study, at a resolution of approximately 9 A, showing that during the incorporation of the aa-tRNA into the 70S ribosome of Escherichia coli, the flexibility of aa-tRNA allows the initial codon recognition and its accommodation into the ribosomal A site. In addition, a conformational change observed in the GTPase-associated center (GAC) of the ribosomal 50S subunit may provide the mechanism by which the ribosome promotes a relative movement of the aa-tRNA with respect to EF-Tu. This relative rearrangement seems to facilitate codon recognition by the incoming aa-tRNA, and to provide the codon-anticodon recognition-dependent signal for the GTPase activity of EF-Tu. From these new findings we propose a mechanism that can explain the sequence of events during the decoding of mRNA on the ribosome. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy.,Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J Nat Struct Biol. 2003 Nov;10(11):899-906. Epub 2003 Oct 19. PMID:14566331[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||