1ne3: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image: | ==Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744== | ||
<StructureSection load='1ne3' size='340' side='right' caption='[[1ne3]], [[NMR_Ensembles_of_Models | 20 NMR models]]' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1ne3]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Methanothermococcus_thermolithotrophicus Methanothermococcus thermolithotrophicus]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1NE3 OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1NE3 FirstGlance]. <br> | |||
</td></tr><tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1ne3 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ne3 OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1ne3 RCSB], [http://www.ebi.ac.uk/pdbsum/1ne3 PDBsum]</span></td></tr> | |||
<table> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ne/1ne3_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
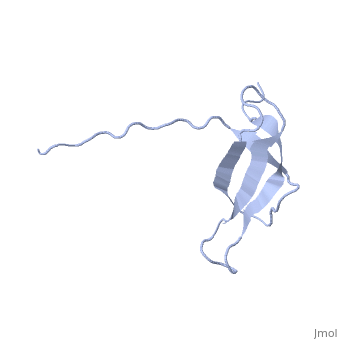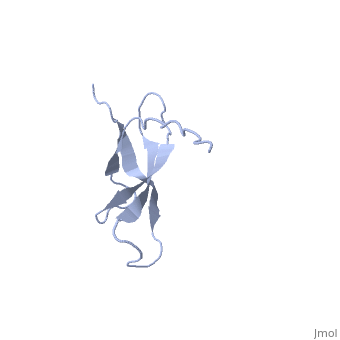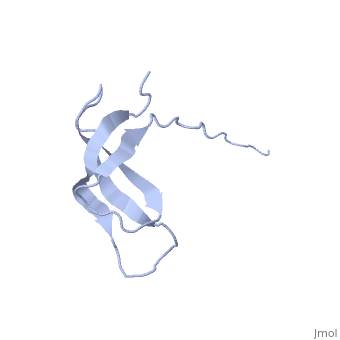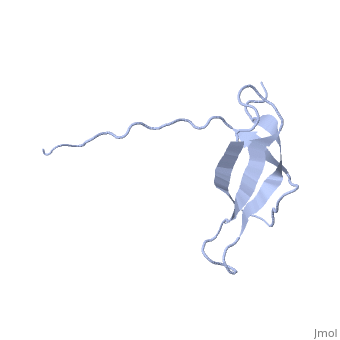
The ribosomal protein S28E from the archaeon Methanobacterium thermoautotrophicum is a component of the 30S ribosomal subunit. Sequence homologs of S28E are found only in archaea and eukaryotes. Here we report the three-dimensional solution structure of S28E by NMR spectroscopy. S28E contains a globular region and a long C-terminal tail protruding from the core. The globular region consists of four antiparallel beta-strands that are arranged in a Greek-key topology. Unique features of S28E include an extended loop L2-3 that folds back onto the protein and a 12-residue charged C-terminal tail with no regular secondary structure and greater flexibility relative to the rest of the protein. The structural and surface resemblance to OB-fold family of proteins and the presence of highly conserved basic residues suggest that S28E may bind to RNA. A broad positively charged surface extending over one side of the beta-barrel and into the flexible C terminus may present a putative binding site for RNA. | |||
Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum.,Wu B, Yee A, Pineda-Lucena A, Semesi A, Ramelot TA, Cort JR, Jung JW, Edwards A, Lee W, Kennedy M, Arrowsmith CH Protein Sci. 2003 Dec;12(12):2831-7. PMID:14627743<ref>PMID:14627743</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
==See Also== | ==See Also== | ||
*[[Ribosomal protein S28|Ribosomal protein S28]] | *[[Ribosomal protein S28|Ribosomal protein S28]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Methanothermococcus thermolithotrophicus]] | [[Category: Methanothermococcus thermolithotrophicus]] | ||
[[Category: Arrowsmith, C H.]] | [[Category: Arrowsmith, C H.]] | ||
Revision as of 17:43, 28 September 2014
Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744Solution structure of ribosomal protein S28E from Methanobacterium Thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH0256_1_68; Northeast Structural Genomics Target TT744
Structural highlights
Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe ribosomal protein S28E from the archaeon Methanobacterium thermoautotrophicum is a component of the 30S ribosomal subunit. Sequence homologs of S28E are found only in archaea and eukaryotes. Here we report the three-dimensional solution structure of S28E by NMR spectroscopy. S28E contains a globular region and a long C-terminal tail protruding from the core. The globular region consists of four antiparallel beta-strands that are arranged in a Greek-key topology. Unique features of S28E include an extended loop L2-3 that folds back onto the protein and a 12-residue charged C-terminal tail with no regular secondary structure and greater flexibility relative to the rest of the protein. The structural and surface resemblance to OB-fold family of proteins and the presence of highly conserved basic residues suggest that S28E may bind to RNA. A broad positively charged surface extending over one side of the beta-barrel and into the flexible C terminus may present a putative binding site for RNA. Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum.,Wu B, Yee A, Pineda-Lucena A, Semesi A, Ramelot TA, Cort JR, Jung JW, Edwards A, Lee W, Kennedy M, Arrowsmith CH Protein Sci. 2003 Dec;12(12):2831-7. PMID:14627743[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences |
| ||||||||||||||