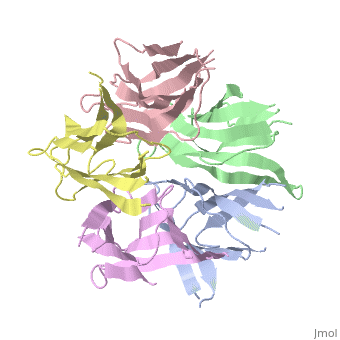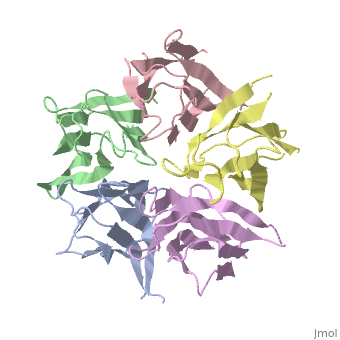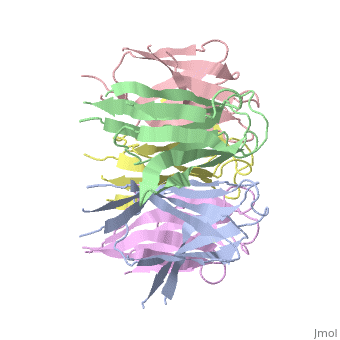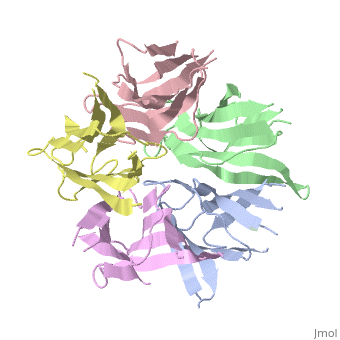1k5j: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image: | ==The Crystal Structure of Nucleoplasmin-Core== | ||
<StructureSection load='1k5j' size='340' side='right' caption='[[1k5j]], [[Resolution|resolution]] 2.30Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1k5j]] is a 5 chain structure with sequence from [http://en.wikipedia.org/wiki/Xenopus_laevis Xenopus laevis]. The January 2007 RCSB PDB [http://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Importins'' by David S. Goodsell is [http://dx.doi.org/10.2210/rcsb_pdb/mom_2007_1 10.2210/rcsb_pdb/mom_2007_1]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1K5J OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1K5J FirstGlance]. <br> | |||
</td></tr><tr><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">Nucleoplasmin ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=8355 Xenopus laevis])</td></tr> | |||
<tr><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1k5j FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1k5j OCA], [http://www.rcsb.org/pdb/explore.do?structureId=1k5j RCSB], [http://www.ebi.ac.uk/pdbsum/1k5j PDBsum]</span></td></tr> | |||
<table> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/k5/1k5j_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/chain_selection.php?pdb_ID=2ata ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
The efficient assembly of histone complexes and nucleosomes requires the participation of molecular chaperones. Currently, there is a paucity of data on their mechanism of action. We now present the structure of an N-terminal domain of nucleoplasmin (Np-core) at 2.3 A resolution. The Np-core monomer is an eight-stranded beta barrel that fits snugly within a stable pentamer. In the crystal, two pentamers associate to form a decamer. We show that both Np and Np-core are competent to assemble large complexes that contain the four core histones. Further experiments and modeling suggest that these complexes each contain five histone octamers which dock to a central Np decamer. This work has important ramifications for models of histone storage, sperm chromatin decondensation, and nucleosome assembly. | |||
The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly.,Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW Mol Cell. 2001 Oct;8(4):841-53. PMID:11684019<ref>PMID:11684019</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
==See Also== | ==See Also== | ||
*[[Nucleoplasmin|Nucleoplasmin]] | *[[Nucleoplasmin|Nucleoplasmin]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Importins]] | [[Category: Importins]] | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
Revision as of 16:32, 28 September 2014
The Crystal Structure of Nucleoplasmin-CoreThe Crystal Structure of Nucleoplasmin-Core
Structural highlights
Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe efficient assembly of histone complexes and nucleosomes requires the participation of molecular chaperones. Currently, there is a paucity of data on their mechanism of action. We now present the structure of an N-terminal domain of nucleoplasmin (Np-core) at 2.3 A resolution. The Np-core monomer is an eight-stranded beta barrel that fits snugly within a stable pentamer. In the crystal, two pentamers associate to form a decamer. We show that both Np and Np-core are competent to assemble large complexes that contain the four core histones. Further experiments and modeling suggest that these complexes each contain five histone octamers which dock to a central Np decamer. This work has important ramifications for models of histone storage, sperm chromatin decondensation, and nucleosome assembly. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly.,Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW Mol Cell. 2001 Oct;8(4):841-53. PMID:11684019[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences |
| ||||||||||||||||